Structural analysis and stochastic modelling suggest a mechanism for calmodulin trapping by CaMKII
- PMID: 22279535
- PMCID: PMC3261145
- DOI: 10.1371/journal.pone.0029406
Structural analysis and stochastic modelling suggest a mechanism for calmodulin trapping by CaMKII
Abstract
Activation of CaMKII by calmodulin and the subsequent maintenance of constitutive activity through autophosphorylation at threonine residue 286 (Thr286) are thought to play a major role in synaptic plasticity. One of the effects of autophosphorylation at Thr286 is to increase the apparent affinity of CaMKII for calmodulin, a phenomenon known as "calmodulin trapping". It has previously been suggested that two binding sites for calmodulin exist on CaMKII, with high and low affinities, respectively. We built structural models of calmodulin bound to both of these sites. Molecular dynamics simulation showed that while binding of calmodulin to the supposed low-affinity binding site on CaMKII is compatible with closing (and hence, inactivation) of the kinase, and could even favour it, binding to the high-affinity site is not. Stochastic simulations of a biochemical model showed that the existence of two such binding sites, one of them accessible only in the active, open conformation, would be sufficient to explain calmodulin trapping by CaMKII. We can explain the effect of CaMKII autophosphorylation at Thr286 on calmodulin trapping: It stabilises the active state and therefore makes the high-affinity binding site accessible. Crucially, a model with only one binding site where calmodulin binding and CaMKII inactivation are strictly mutually exclusive cannot reproduce calmodulin trapping. One of the predictions of our study is that calmodulin binding in itself is not sufficient for CaMKII activation, although high-affinity binding of calmodulin is.
Conflict of interest statement
Figures






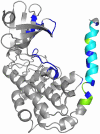
 Å cutoff.
Å cutoff.
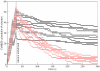
 . The ratio of calmodulin to CaMKII concentration used in the simulation was the same as used in the experimental setup by Meyer et al. . The number of calmodulin-bound monomeric CaMKII subunits is plotted against time for each simulation run. The total number of CaMKII subunits in the simulation was 360. Wildtype is shown in black, T286A mutant in red. Ten simulation runs are shown for each.
. The ratio of calmodulin to CaMKII concentration used in the simulation was the same as used in the experimental setup by Meyer et al. . The number of calmodulin-bound monomeric CaMKII subunits is plotted against time for each simulation run. The total number of CaMKII subunits in the simulation was 360. Wildtype is shown in black, T286A mutant in red. Ten simulation runs are shown for each.
 to
to  . The three levels in the y-dimension represent calmodulin binding, with no binding (lowest level), low-affinity binding (middle level) and high-affinity binding (highest level). Events of calmodulin sliding back and forth between the high-affinity and the low-affinity binding sites appear as drops from the top level to the centre and back up. The colour of the trace represents subunit phosphorylation at Thr286, with unphosphorylated subunits shown in black, and phosphorylated subunits in red.
. The three levels in the y-dimension represent calmodulin binding, with no binding (lowest level), low-affinity binding (middle level) and high-affinity binding (highest level). Events of calmodulin sliding back and forth between the high-affinity and the low-affinity binding sites appear as drops from the top level to the centre and back up. The colour of the trace represents subunit phosphorylation at Thr286, with unphosphorylated subunits shown in black, and phosphorylated subunits in red.
Similar articles
-
A multi-state model of the CaMKII dodecamer suggests a role for calmodulin in maintenance of autophosphorylation.PLoS Comput Biol. 2019 Dec 23;15(12):e1006941. doi: 10.1371/journal.pcbi.1006941. eCollection 2019 Dec. PLoS Comput Biol. 2019. PMID: 31869343 Free PMC article.
-
Substrate-selective and calcium-independent activation of CaMKII by α-actinin.J Biol Chem. 2012 May 4;287(19):15275-83. doi: 10.1074/jbc.M112.351817. Epub 2012 Mar 15. J Biol Chem. 2012. PMID: 22427672 Free PMC article.
-
Conformational changes underlying calcium/calmodulin-dependent protein kinase II activation.EMBO J. 2011 Apr 6;30(7):1251-62. doi: 10.1038/emboj.2011.40. Epub 2011 Feb 22. EMBO J. 2011. PMID: 21343908 Free PMC article.
-
Regulation and role of brain calcium/calmodulin-dependent protein kinase II.Neurochem Int. 1992 Dec;21(4):469-97. doi: 10.1016/0197-0186(92)90080-b. Neurochem Int. 1992. PMID: 1338943 Review.
-
Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function.Annu Rev Biochem. 2002;71:473-510. doi: 10.1146/annurev.biochem.71.110601.135410. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045104 Review.
Cited by
-
Calcium input frequency, duration and amplitude differentially modulate the relative activation of calcineurin and CaMKII.PLoS One. 2012;7(9):e43810. doi: 10.1371/journal.pone.0043810. Epub 2012 Sep 4. PLoS One. 2012. PMID: 22962589 Free PMC article.
-
Biophysical attributes that affect CaMKII activation deduced with a novel spatial stochastic simulation approach.PLoS Comput Biol. 2018 Feb 5;14(2):e1005946. doi: 10.1371/journal.pcbi.1005946. eCollection 2018 Feb. PLoS Comput Biol. 2018. PMID: 29401454 Free PMC article.
-
Real-time single-molecule imaging of CaMKII-calmodulin interactions.Biophys J. 2024 Apr 2;123(7):824-838. doi: 10.1016/j.bpj.2024.02.021. Epub 2024 Feb 28. Biophys J. 2024. PMID: 38414237 Free PMC article.
-
Validation Through Collaboration: Encouraging Team Efforts to Ensure Internal and External Validity of Computational Models of Biochemical Pathways.Neuroinformatics. 2022 Jan;20(1):277-284. doi: 10.1007/s12021-022-09584-5. Epub 2022 May 11. Neuroinformatics. 2022. PMID: 35543917 Free PMC article. Review.
-
The Effect of Ca2+, Lobe-Specificity, and CaMKII on CaM Binding to NaV1.1.Int J Mol Sci. 2018 Aug 23;19(9):2495. doi: 10.3390/ijms19092495. Int J Mol Sci. 2018. PMID: 30142967 Free PMC article.
References
-
- Bennett MK, Erondu NE, Kennedy MB. Purification and characterization of a calmodulindependent protein kinase that is highly concentrated in brain. J Biol Chem. 1983;258:12735–12744. - PubMed
-
- Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calciumcalmodulin kinase II mutant mice. Science. 1992;257:206–211. - PubMed
-
- Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. - PubMed
-
- Whitlock JR, Heynen AJ, Shuler MG, Bear MF. Learning induces long-term potentiation in the hippocampus. Science. 2006;313:1093–1097. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

