Genetic signatures of exceptional longevity in humans
- PMID: 22279548
- PMCID: PMC3261167
- DOI: 10.1371/journal.pone.0029848
Genetic signatures of exceptional longevity in humans
Abstract
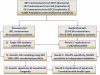
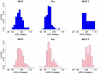
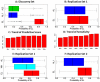
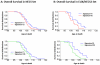
Like most complex phenotypes, exceptional longevity is thought to reflect a combined influence of environmental (e.g., lifestyle choices, where we live) and genetic factors. To explore the genetic contribution, we undertook a genome-wide association study of exceptional longevity in 801 centenarians (median age at death 104 years) and 914 genetically matched healthy controls. Using these data, we built a genetic model that includes 281 single nucleotide polymorphisms (SNPs) and discriminated between cases and controls of the discovery set with 89% sensitivity and specificity, and with 58% specificity and 60% sensitivity in an independent cohort of 341 controls and 253 genetically matched nonagenarians and centenarians (median age 100 years). Consistent with the hypothesis that the genetic contribution is largest with the oldest ages, the sensitivity of the model increased in the independent cohort with older and older ages (71% to classify subjects with an age at death>102 and 85% to classify subjects with an age at death>105). For further validation, we applied the model to an additional, unmatched 60 centenarians (median age 107 years) resulting in 78% sensitivity, and 2863 unmatched controls with 61% specificity. The 281 SNPs include the SNP rs2075650 in TOMM40/APOE that reached irrefutable genome wide significance (posterior probability of association = 1) and replicated in the independent cohort. Removal of this SNP from the model reduced the accuracy by only 1%. Further in-silico analysis suggests that 90% of centenarians can be grouped into clusters characterized by different "genetic signatures" of varying predictive values for exceptional longevity. The correlation between 3 signatures and 3 different life spans was replicated in the combined replication sets. The different signatures may help dissect this complex phenotype into sub-phenotypes of exceptional longevity.
Conflict of interest statement
Figures






References
-
- Fraser GE, Shavlik DJ. Ten years of life: Is it a matter of choice? Arch Intern Med. 2001;161:1645–1652. - PubMed
-
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. - PubMed
-
- Alpert L, DesJardines B, Vaupel J, Perls Tt. Extreme longevity in two families. A report of multiple centenarians within single generations. In: Jeune BVJ, editor. Age Validation of the Extreme Old. Odense: Odense University Press; 1998.
-
- Perls T, Shea-Drinkwater M, Bowen-Flynn J, Ridge SB, Kang S, et al. Exceptional familial clustering for extreme longevity in humans. J Am Geriatr Soc. 2000;48:1483–1485. - PubMed
-
- Westendorp RG, van Heemst D, Rozing MP, Frolich M, Mooijaart SP, et al. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: The Leiden Longevity Study. J Am Geriatr Soc. 2009;57:1634–1637. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

