Sequence coevolution between RNA and protein characterized by mutual information between residue triplets
- PMID: 22279560
- PMCID: PMC3261191
- DOI: 10.1371/journal.pone.0030022
Sequence coevolution between RNA and protein characterized by mutual information between residue triplets
Abstract
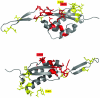
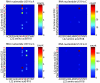
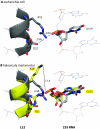
Coevolving residues in a multiple sequence alignment provide evolutionary clues of biophysical interactions in 3D structure. Despite a rich literature describing amino acid coevolution within or between proteins and nucleic acid coevolution within RNA, to date there has been no direct evidence of coevolution between protein and RNA. The ribosome, a structurally conserved macromolecular machine composed of over 50 interacting protein and RNA chains, provides a natural example of RNA/protein interactions that likely coevolved. We provide the first direct evidence of RNA/protein coevolution by characterizing the mutual information in residue triplets from a multiple sequence alignment of ribosomal protein L22 and neighboring 23S RNA. We define residue triplets as three positions in the multiple sequence alignment, where one position is from the 23S RNA and two positions are from the L22 protein. We show that residue triplets with high mutual information are more likely than residue doublets to be proximal in 3D space. Some high mutual information residue triplets cluster in a connected series across the L22 protein structure, similar to patterns seen in protein coevolution. We also describe RNA nucleotides for which switching from one nucleotide to another (or between purines and pyrimidines) results in a change in amino acid distribution for proximal amino acid positions. Multiple crystal structures for evolutionarily distinct ribosome species can provide structural evidence for these differences. For one residue triplet, a pyrimidine in one species is a purine in another, and RNA/protein hydrogen bonds are present in one species but not the other. The results provide the first direct evidence of RNA/protein coevolution by using higher order mutual information, suggesting that biophysical constraints on interacting RNA and protein chains are indeed a driving force in their evolution.
Conflict of interest statement
Figures

Similar articles
-
Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions.Biochemistry. 2005 May 17;44(19):7156-65. doi: 10.1021/bi050293e. Biochemistry. 2005. PMID: 15882054
-
CoeViz: a web-based tool for coevolution analysis of protein residues.BMC Bioinformatics. 2016 Mar 8;17:119. doi: 10.1186/s12859-016-0975-z. BMC Bioinformatics. 2016. PMID: 26956673 Free PMC article.
-
Protein-protein interactions leave evolutionary footprints: High molecular coevolution at the core of interfaces.Protein Sci. 2017 Dec;26(12):2438-2444. doi: 10.1002/pro.3318. Epub 2017 Oct 25. Protein Sci. 2017. PMID: 28980349 Free PMC article.
-
Structure and dynamics of ribosomal RNA.Curr Opin Struct Biol. 1998 Jun;8(3):294-300. doi: 10.1016/s0959-440x(98)80061-4. Curr Opin Struct Biol. 1998. PMID: 9666324 Review.
-
RNA-amino acid binding: a stereochemical era for the genetic code.J Mol Evol. 2009 Nov;69(5):406-29. doi: 10.1007/s00239-009-9270-1. Epub 2009 Oct 1. J Mol Evol. 2009. PMID: 19795157 Review.
Cited by
-
Impact of human pathogenic micro-insertions and micro-deletions on post-transcriptional regulation.Hum Mol Genet. 2014 Jun 1;23(11):3024-34. doi: 10.1093/hmg/ddu019. Epub 2014 Jan 16. Hum Mol Genet. 2014. PMID: 24436305 Free PMC article.
-
In the Beginning was a Mutualism - On the Origin of Translation.Orig Life Evol Biosph. 2018 Jun;48(2):223-243. doi: 10.1007/s11084-018-9557-6. Epub 2018 Apr 30. Orig Life Evol Biosph. 2018. PMID: 29713988
-
Evolution of the RNA alternative decay cis element into a high-affinity target for the immunomodulatory protein Roquin.RNA Biol. 2025 Dec;22(1):1-12. doi: 10.1080/15476286.2024.2448391. Epub 2025 Jan 13. RNA Biol. 2025. PMID: 39801464 Free PMC article.
-
Shaping the regulation of the p53 mRNA tumour suppressor: the co-evolution of genetic signatures.BMC Cancer. 2019 Sep 13;19(1):915. doi: 10.1186/s12885-019-6118-y. BMC Cancer. 2019. PMID: 31519161 Free PMC article. Review.
-
Imp/IGF2BP and Syp/SYNCRIP temporal RNA interactomes uncover combinatorial networks of regulators of Drosophila brain development.Sci Adv. 2025 Feb 7;11(6):eadr6682. doi: 10.1126/sciadv.adr6682. Epub 2025 Feb 7. Sci Adv. 2025. PMID: 39919181 Free PMC article.
References
-
- Fitch WM, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970;4:579–593. - PubMed
-
- Yanofsky C, Horn V, Thorpe D. Protein Structure Relationships Revealed by Mutational Analysis. Science. 1964;146:1593–1594. - PubMed
-
- Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

