ArfGAP1 promotes COPI vesicle formation by facilitating coatomer polymerization
- PMID: 22279613
- PMCID: PMC3265926
- DOI: 10.4161/cl.1.4.18896
ArfGAP1 promotes COPI vesicle formation by facilitating coatomer polymerization
Abstract
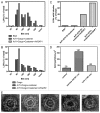
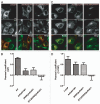
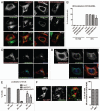
The role of ArfGAP1 in COPI vesicle biogenesis has been controversial. In work using isolated Golgi membranes, ArfGAP1 was found to promote COPI vesicle formation. In contrast, in studies using large unilamellar vesicles (LUVs) as model membranes, ArfGAP1 functioned as an uncoating factor inhibiting COPI vesicle formation. We set out to discriminate between these models. First, we reexamined the effect of ArfGAP1 on LUVs. We found that ArfGAP1 increased the efficiency of coatomer-induced deformation of LUVs. Second, ArfGAP1 and peptides from cargo facilitated self-assembly of coatomer into spherical structures in the absence of membranes, reminiscent of clathrin self-assembly. Third, in vivo, ArfGAP1 overexpression induced the accumulation of vesicles and allowed normal trafficking of a COPI cargo. Taken together, these data support the model in which ArfGAP1 promotes COPI vesicle formation and membrane traffic and identify a function for ArfGAP1 in the assembly of coatomer into COPI.
Keywords: ADP-ribosylation factor; ArfGTPase-activating protein; Golgi apparatus; coatomer; membrane traffic.
Figures






References
LinkOut - more resources
Full Text Sources
