Evolutionarily assembled cis-regulatory module at a human ciliopathy locus
- PMID: 22282472
- PMCID: PMC3671610
- DOI: 10.1126/science.1213506
Evolutionarily assembled cis-regulatory module at a human ciliopathy locus
Abstract
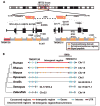
Neighboring genes are often coordinately expressed within cis-regulatory modules, but evidence that nonparalogous genes share functions in mammals is lacking. Here, we report that mutation of either TMEM138 or TMEM216 causes a phenotypically indistinguishable human ciliopathy, Joubert syndrome. Despite a lack of sequence homology, the genes are aligned in a head-to-tail configuration and joined by chromosomal rearrangement at the amphibian-to-reptile evolutionary transition. Expression of the two genes is mediated by a conserved regulatory element in the noncoding intergenic region. Coordinated expression is important for their interdependent cellular role in vesicular transport to primary cilia. Hence, during vertebrate evolution of genes involved in ciliogenesis, nonparalogous genes were arranged to a functional gene cluster with shared regulatory elements.
Figures



Comment in
-
Genetics. Mendelian puzzles.Science. 2012 Feb 24;335(6071):930-1. doi: 10.1126/science.1219301. Science. 2012. PMID: 22362999 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- R01 EY021872/EY/NEI NIH HHS/United States
- DK068306/DK/NIDDK NIH HHS/United States
- DK072301/DK/NIDDK NIH HHS/United States
- NS052455/NS/NINDS NIH HHS/United States
- HD042601/HD/NICHD NIH HHS/United States
- R01 NS052455/NS/NINDS NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- DK075972/DK/NIDDK NIH HHS/United States
- NS04843/NS/NINDS NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P30NS047101/NS/NINDS NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- R01 DK072301/DK/NIDDK NIH HHS/United States
- R01 NS048453/NS/NINDS NIH HHS/United States
- G0700073/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- R01 DK068306/DK/NIDDK NIH HHS/United States
- EY021872/EY/NEI NIH HHS/United States
- GGP08145/TI_/Telethon/Italy
- R01 DK075972/DK/NIDDK NIH HHS/United States
- DK090917/DK/NIDDK NIH HHS/United States
- RC4 DK090917/DK/NIDDK NIH HHS/United States
- R01 HD042601/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

