Repeatability and contingency in the evolution of a key innovation in phage lambda
- PMID: 22282803
- PMCID: PMC3306806
- DOI: 10.1126/science.1214449
Repeatability and contingency in the evolution of a key innovation in phage lambda
Abstract
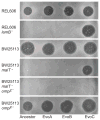
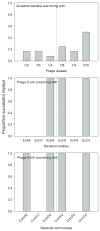
The processes responsible for the evolution of key innovations, whereby lineages acquire qualitatively new functions that expand their ecological opportunities, remain poorly understood. We examined how a virus, bacteriophage λ, evolved to infect its host, Escherichia coli, through a novel pathway. Natural selection promoted the fixation of mutations in the virus's host-recognition protein, J, that improved fitness on the original receptor, LamB, and set the stage for other mutations that allowed infection through a new receptor, OmpF. These viral mutations arose after the host evolved reduced expression of LamB, whereas certain other host mutations prevented the phage from evolving the new function. This study shows the complex interplay between genomic processes and ecological conditions that favor the emergence of evolutionary innovations.
Figures



Comment in
-
Evolution. The role of coevolution.Science. 2012 Jan 27;335(6067):410-1. doi: 10.1126/science.1217807. Science. 2012. PMID: 22282796 No abstract available.
References
-
- Mayr E. Animal Species and Evolution. Belknap Press; Cambridge, MA: 1963.
-
- Gavrilets S. In: Toward an Extended Evolutionary Synthesis. Pigliucci M, Muller G, editors. MIT Press; Cambridge, MA: 2010.
-
- Darwin C. The Origin of Species. Murray; London: 1859.
-
- Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press; Oxford: 1930.
-
- Dieckmann U, Doebeli M, Metz JAJ, editors. Adaptive Speciation. Cambridge Univ. Press; Cambridge, UK: 2004.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

