Two ZnF-UBP domains in isopeptidase T (USP5)
- PMID: 22283393
- PMCID: PMC8391072
- DOI: 10.1021/bi200854q
Two ZnF-UBP domains in isopeptidase T (USP5)
Abstract
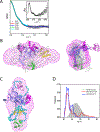
Human ubiquitin-specific cysteine protease 5 (USP5, also known as ISOT and isopeptidase T), an 835-residue multidomain enzyme, recycles ubiquitin by hydrolyzing isopeptide bonds in a variety of unanchored polyubiquitin substrates. Activation of the enzyme's hydrolytic activity toward ubiquitin-AMC (7-amino-4-methylcoumarin), a fluorogenic substrate, by the addition of free, unanchored monoubiquitin suggested an allosteric mechanism of activation by the ZnF-UBP domain (residues 163-291), which binds the substrate's unanchored diglycine carboxyl tail. By determining the structure of full-length USP5, we discovered the existence of a cryptic ZnF-UBP domain (residues 1-156), which was tightly bound to the catalytic core and was indispensable for catalytic activity. In contrast, the previously characterized ZnF-UBP domain did not contribute directly to the active site; a paucity of interactions suggested flexibility between these two domains consistent with an ability by the enzyme to hydrolyze a variety of different polyubiquitin chain linkages. Deletion of the known ZnF-UBP domain did not significantly affect rate of hydrolysis of ubiquitin-AMC and suggested that it is likely associated mainly with substrate targeting and specificity. Together, our findings show that USP5 uses multiple ZnF-UBP domains for substrate targeting and core catalytic function.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Figures
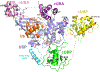


References
-
- Komander D (2009) The emerging complexity of protein ubiquitination, Biochem Soc Trans 37, 937–953. - PubMed
-
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, and Gygi SP (2003) A proteomics approach to understanding protein ubiquitination, Nat Biotechnol 21, 921–926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

