Genome-wide association studies for agronomical traits in a world wide spring barley collection
- PMID: 22284310
- PMCID: PMC3349577
- DOI: 10.1186/1471-2229-12-16
Genome-wide association studies for agronomical traits in a world wide spring barley collection
Abstract
Background: Genome-wide association studies (GWAS) based on linkage disequilibrium (LD) provide a promising tool for the detection and fine mapping of quantitative trait loci (QTL) underlying complex agronomic traits. In this study we explored the genetic basis of variation for the traits heading date, plant height, thousand grain weight, starch content and crude protein content in a diverse collection of 224 spring barleys of worldwide origin. The whole panel was genotyped with a customized oligonucleotide pool assay containing 1536 SNPs using Illumina's GoldenGate technology resulting in 957 successful SNPs covering all chromosomes. The morphological trait "row type" (two-rowed spike vs. six-rowed spike) was used to confirm the high level of selectivity and sensitivity of the approach. This study describes the detection of QTL for the above mentioned agronomic traits by GWAS.
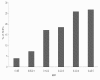
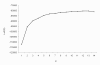
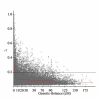
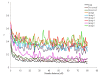
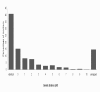
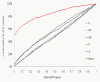
Results: Population structure in the panel was investigated by various methods and six subgroups that are mainly based on their spike morphology and region of origin. We explored the patterns of linkage disequilibrium (LD) among the whole panel for all seven barley chromosomes. Average LD was observed to decay below a critical level (r2-value 0.2) within a map distance of 5-10 cM. Phenotypic variation within the panel was reasonably large for all the traits. The heritabilities calculated for each trait over multi-environment experiments ranged between 0.90-0.95. Different statistical models were tested to control spurious LD caused by population structure and to calculate the P-value of marker-trait associations. Using a mixed linear model with kinship for controlling spurious LD effects, we found a total of 171 significant marker trait associations, which delineate into 107 QTL regions. Across all traits these can be grouped into 57 novel QTL and 50 QTL that are congruent with previously mapped QTL positions.
Conclusions: Our results demonstrate that the described diverse barley panel can be efficiently used for GWAS of various quantitative traits, provided that population structure is appropriately taken into account. The observed significant marker trait associations provide a refined insight into the genetic architecture of important agronomic traits in barley. However, individual QTL account only for a small portion of phenotypic variation, which may be due to insufficient marker coverage and/or the elimination of rare alleles prior to analysis. The fact that the combined SNP effects fall short of explaining the complete phenotypic variance may support the hypothesis that the expression of a quantitative trait is caused by a large number of very small effects that escape detection. Notwithstanding these limitations, the integration of GWAS with biparental linkage mapping and an ever increasing body of genomic sequence information will facilitate the systematic isolation of agronomically important genes and subsequent analysis of their allelic diversity.
Figures

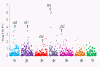


Similar articles
-
DNA polymorphisms and haplotype patterns of transcription factors involved in barley endosperm development are associated with key agronomic traits.BMC Plant Biol. 2010 Jan 8;10:5. doi: 10.1186/1471-2229-10-5. BMC Plant Biol. 2010. PMID: 20064201 Free PMC article.
-
Marker-trait associations in Virginia Tech winter barley identified using genome-wide mapping.Theor Appl Genet. 2013 Mar;126(3):693-710. doi: 10.1007/s00122-012-2011-7. Epub 2012 Nov 9. Theor Appl Genet. 2013. PMID: 23139143
-
Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.).BMC Genet. 2006 Jan 24;7:6. doi: 10.1186/1471-2156-7-6. BMC Genet. 2006. PMID: 16433922 Free PMC article.
-
GWAS for identification of genomic regions and candidate genes in vegetable crops.Funct Integr Genomics. 2024 Oct 29;24(6):203. doi: 10.1007/s10142-024-01477-x. Funct Integr Genomics. 2024. PMID: 39470821 Review.
-
GWAS: Fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley - A review.J Adv Res. 2019 Nov 4;22:119-135. doi: 10.1016/j.jare.2019.10.013. eCollection 2020 Mar. J Adv Res. 2019. PMID: 31956447 Free PMC article. Review.
Cited by
-
Genome-wide association mapping of salinity tolerance in rice (Oryza sativa).DNA Res. 2015 Apr;22(2):133-45. doi: 10.1093/dnares/dsu046. Epub 2015 Jan 27. DNA Res. 2015. PMID: 25627243 Free PMC article.
-
Genome-wide association analysis of grain yield and Striga hermonthica and S. asiatica resistance in tropical and sub-tropical maize populations.BMC Plant Biol. 2024 Sep 19;24(1):871. doi: 10.1186/s12870-024-05590-8. BMC Plant Biol. 2024. PMID: 39294608 Free PMC article.
-
SNP-Based QTL Mapping of 15 Complex Traits in Barley under Rain-Fed and Well-Watered Conditions by a Mixed Modeling Approach.Front Plant Sci. 2016 Jun 27;7:909. doi: 10.3389/fpls.2016.00909. eCollection 2016. Front Plant Sci. 2016. PMID: 27446139 Free PMC article.
-
Association mapping for agronomic traits in six-rowed spring barley from the USA harvested in Kazakhstan.PLoS One. 2019 Aug 12;14(8):e0221064. doi: 10.1371/journal.pone.0221064. eCollection 2019. PLoS One. 2019. PMID: 31404111 Free PMC article.
-
Association Mapping of Amylose Content in Maize RIL Population Using SSR and SNP Markers.Plants (Basel). 2023 Jan 4;12(2):239. doi: 10.3390/plants12020239. Plants (Basel). 2023. PMID: 36678952 Free PMC article.
References
-
- Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. - PubMed
-
- Zhu C, Gore M, Buckler ES, Yu J. Status and Prospects of Association Mapping in Plants. The Plant Genome Journal. 2008;1(1):5.
-
- Hastbacka J, Delachapelle A, Kaitila I, Sistonen P, Weaver A, Lander E. Linkage Disequilibrium Mapping in Isolated Founder Populations - Diastrophic Dysplasia in Finland. Nature Genetics. 1992;2(3):204–211. - PubMed
-
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–2048. - PubMed
-
- Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44(6):1054–1064. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

