DNA base excision repair: a mechanism of trinucleotide repeat expansion
- PMID: 22285516
- PMCID: PMC3323758
- DOI: 10.1016/j.tibs.2011.12.002
DNA base excision repair: a mechanism of trinucleotide repeat expansion
Abstract
The expansion of trinucleotide repeat (TNR) sequences in human DNA is considered to be a key factor in the pathogenesis of more than 40 neurodegenerative diseases. TNR expansion occurs during DNA replication and also, as suggested by recent studies, during the repair of DNA lesions produced by oxidative stress. In particular, the oxidized guanine base 8-oxoguanine within sequences containing CAG repeats may induce formation of pro-expansion intermediates through strand slippage during DNA base excision repair (BER). In this article, we describe how oxidized DNA lesions are repaired by BER and discuss the importance of the coordinated activities of the key repair enzymes, such as DNA polymerase β, flap endonuclease 1 (FEN1) and DNA ligase, in preventing strand slippage and TNR expansion.
Published by Elsevier Ltd.
Figures
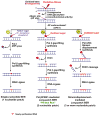
 ) of Okazaki fragments. The flaps are created by pol δ or pol ε strand-displacement synthesis. FEN1 usually captures a dual flap intermediate with a one-nucleotide 3′-flap along with a 5′-flap and removes the 5′-flap, leaving a nicked DNA that is a substrate for ligation, a “ligatable nick.” (b) During long-patch BER, FEN1 removes a modified (reduced or oxidized) sugar (
) of Okazaki fragments. The flaps are created by pol δ or pol ε strand-displacement synthesis. FEN1 usually captures a dual flap intermediate with a one-nucleotide 3′-flap along with a 5′-flap and removes the 5′-flap, leaving a nicked DNA that is a substrate for ligation, a “ligatable nick.” (b) During long-patch BER, FEN1 removes a modified (reduced or oxidized) sugar (
 ) by either coordinating with pol β through the “Hit and Run” mechanism (the sub-pathway on the left) or capturing a 5′-flap associated with a modified sugar (the sub-pathway on the right) created by pol β or pol δ/ε. This latter process results in multi-nucleotide replacement. In the case where FEN1 cleavage results in a one-nucleotide gap, pol β will fill the gap readily leaving a ligatable nick.
) by either coordinating with pol β through the “Hit and Run” mechanism (the sub-pathway on the left) or capturing a 5′-flap associated with a modified sugar (the sub-pathway on the right) created by pol β or pol δ/ε. This latter process results in multi-nucleotide replacement. In the case where FEN1 cleavage results in a one-nucleotide gap, pol β will fill the gap readily leaving a ligatable nick.
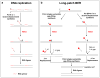
 ) cannot be removed by pol β’s dRP lyase. Strand slippage results in formation of intermediates with multi-nucleotide gaps and repeat-containing hairpins. Pol β conducts gap-filling synthesis to fill the gaps, but terminates synthesis at the base of the hairpin. Formation of a stable hairpin structure inhibits conventional FEN1 cleavage activity for removing the entire length of the hairpin. Instead, FEN1 “alternate cleavage” of a short flap generated at the 5′-end of the hairpin by DNA realignment is allowed, resulting in removal of the flap with the 5′-end dRP group, a ligatable nick, ligation and repeat expansion.
) cannot be removed by pol β’s dRP lyase. Strand slippage results in formation of intermediates with multi-nucleotide gaps and repeat-containing hairpins. Pol β conducts gap-filling synthesis to fill the gaps, but terminates synthesis at the base of the hairpin. Formation of a stable hairpin structure inhibits conventional FEN1 cleavage activity for removing the entire length of the hairpin. Instead, FEN1 “alternate cleavage” of a short flap generated at the 5′-end of the hairpin by DNA realignment is allowed, resulting in removal of the flap with the 5′-end dRP group, a ligatable nick, ligation and repeat expansion.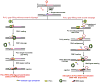
 ) plus 1 nucleotide, pol β then conducts gap-filling (dCMP insertion) and a nicked DNA intermediate is produced. This process occurs without strand slippage that could result in hairpin structures. The nicked intermediate is subsequently sealed by DNA ligase, and repeat expansion will not occur. (Pathway 2) If pol β and FEN1 coordination is disrupted by spontaneous strand slippage, CAG repeat hairpin structures will form, the multi-nucleotide gap will be filled by pol β, and pol β terminates synthesis at the base of the hairpin. FEN1 flap cleavage activity also will be inhibited. Thus, stabile hairpin structures inhibit FEN1 conventional cleavage at the 3′-base of the hairpin, as illustrated. Alternatively, FEN1 “alternate cleavage” of a short 5′-flap at the base of the hairpin will result in a ligatable nick, ligation and repeat expansion.
) plus 1 nucleotide, pol β then conducts gap-filling (dCMP insertion) and a nicked DNA intermediate is produced. This process occurs without strand slippage that could result in hairpin structures. The nicked intermediate is subsequently sealed by DNA ligase, and repeat expansion will not occur. (Pathway 2) If pol β and FEN1 coordination is disrupted by spontaneous strand slippage, CAG repeat hairpin structures will form, the multi-nucleotide gap will be filled by pol β, and pol β terminates synthesis at the base of the hairpin. FEN1 flap cleavage activity also will be inhibited. Thus, stabile hairpin structures inhibit FEN1 conventional cleavage at the 3′-base of the hairpin, as illustrated. Alternatively, FEN1 “alternate cleavage” of a short 5′-flap at the base of the hairpin will result in a ligatable nick, ligation and repeat expansion.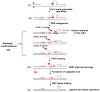
 ) may undergo spontaneous DNA slippage resulting in formation of multi-nucleotide gaps and a CAG repeat-containing hairpin with 5′-sugar phosphate (i). Pol β performs multi-nucleotide gap-filling synthesis to produce a newly synthesized CAG repeat strand, eventually colliding with the 5′-base of the hairpin. Pol β pauses at the base of the hairpin and dissociates from this product (ii). The CAG repeat hairpin undergoes spontaneous DNA realignment. This leads to formation of intermediates with alternate conformations (iii) of an annealed 5′-region consisting of 3 nucleotides (one CAG repeat) and various lengths of 5′-repeat flap, consisting of 3 nt [(CAG)1], 6 nt [(CAG)2], 9 nt [(CAG)3], etc. FEN1 loading and alternate cleavage (iv) then removes the 5′-CAG repeat-containing flaps, creating the ligatable nick for ligation, resulting in TNR expansion (v). During the spontaneous DNA realignment and formation of alternate conformations proposed, the hairpin migrates to the right, as illustrated by the arrow.
) may undergo spontaneous DNA slippage resulting in formation of multi-nucleotide gaps and a CAG repeat-containing hairpin with 5′-sugar phosphate (i). Pol β performs multi-nucleotide gap-filling synthesis to produce a newly synthesized CAG repeat strand, eventually colliding with the 5′-base of the hairpin. Pol β pauses at the base of the hairpin and dissociates from this product (ii). The CAG repeat hairpin undergoes spontaneous DNA realignment. This leads to formation of intermediates with alternate conformations (iii) of an annealed 5′-region consisting of 3 nucleotides (one CAG repeat) and various lengths of 5′-repeat flap, consisting of 3 nt [(CAG)1], 6 nt [(CAG)2], 9 nt [(CAG)3], etc. FEN1 loading and alternate cleavage (iv) then removes the 5′-CAG repeat-containing flaps, creating the ligatable nick for ligation, resulting in TNR expansion (v). During the spontaneous DNA realignment and formation of alternate conformations proposed, the hairpin migrates to the right, as illustrated by the arrow.References
-
- Paulson HL, Fischbeck KH. Trinucleotide repeats in neurogenetic disorders. Annu Rev Neurosci. 1996;19:79–107. - PubMed
-
- Pearson CE, et al. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. - PubMed
-
- Kennedy L, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

