Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments
- PMID: 22286036
- PMCID: PMC3290041
- DOI: 10.1159/000334611
Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments
Abstract
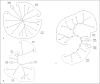
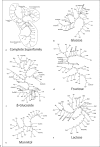
Transport proteins function in the translocation of ions, solutes and macromolecules across cellular and organellar membranes. These integral membrane proteins fall into >600 families as tabulated in the Transporter Classification Database (www.tcdb.org). Recent studies, some of which are reported here, define distant phylogenetic relationships between families with the creation of superfamilies. Several of these are analyzed using a novel set of programs designed to allow reliable prediction of phylogenetic trees when sequence divergence is too great to allow the use of multiple alignments. These new programs, called SuperfamilyTree1 and 2 (SFT1 and 2), allow display of protein and family relationships, respectively, based on thousands of comparative BLAST scores rather than multiple alignments. Superfamilies analyzed include: (1) Aerolysins, (2) RTX Toxins, (3) Defensins, (4) Ion Transporters, (5) Bile/Arsenite/Riboflavin Transporters, (6) Cation:Proton Antiporters, and (7) the Glucose/Fructose/Lactose superfamily within the prokaryotic phosphoenol pyruvate-dependent Phosphotransferase System. In addition to defining the phylogenetic relationships of the proteins and families within these seven superfamilies, evidence is provided showing that the SFT programs outperform programs that are based on multiple alignments whenever sequence divergence of superfamily members is extensive. The SFT programs should be applicable to virtually any superfamily of proteins or nucleic acids.
Copyright © 2012 S. Karger AG, Basel.
Figures





References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- An JH, Kim YS. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii–cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem. 1998;257:395–402. - PubMed
-
- Bhattacharjee H, Zhou T, Li J, Gatti DL, Walmsley AR, Rosen BP. Structure-function relationships in an anion-translocating ATPase. Biochem Soc Trans. 2000;28:520–526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

