Dynamic localization of Trypanosoma brucei mitochondrial DNA polymerase ID
- PMID: 22286095
- PMCID: PMC3416495
- DOI: 10.1128/EC.05291-11
Dynamic localization of Trypanosoma brucei mitochondrial DNA polymerase ID
Abstract
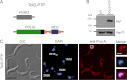
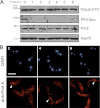
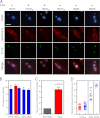
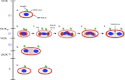
Trypanosomes contain a unique form of mitochondrial DNA called kinetoplast DNA (kDNA) that is a catenated network composed of minicircles and maxicircles. Several proteins are essential for network replication, and most of these localize to the antipodal sites or the kinetoflagellar zone. Essential components for kDNA synthesis include three mitochondrial DNA polymerases TbPOLIB, TbPOLIC, and TbPOLID). In contrast to other kDNA replication proteins, TbPOLID was previously reported to localize throughout the mitochondrial matrix. This spatial distribution suggests that TbPOLID requires redistribution to engage in kDNA replication. Here, we characterize the subcellular distribution of TbPOLID with respect to the Trypanosoma brucei cell cycle using immunofluorescence microscopy. Our analyses demonstrate that in addition to the previously reported matrix localization, TbPOLID was detected as discrete foci near the kDNA. TbPOLID foci colocalized with replicating minicircles at antipodal sites in a specific subset of the cells during stages II and III of kDNA replication. Additionally, the TbPOLID foci were stable following the inhibition of protein synthesis, detergent extraction, and DNase treatment. Taken together, these data demonstrate that TbPOLID has a dynamic localization that allows it to be spatially and temporally available to perform its role in kDNA replication.
Figures


References
-
- Bogenhagen DF, Rousseau D, Burke S. 2008. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283:3665–3675 - PubMed
-
- Bruhn DF, Mozeleski B, Falkin L, Klingbeil MM. 2010. Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes. Mol. Microbiol. 75:1414–1425 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

