Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming
- PMID: 22286172
- PMCID: PMC3315209
- DOI: 10.1093/hmg/dds023
Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming
Abstract
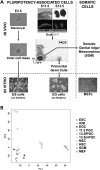
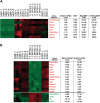
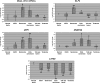
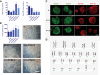
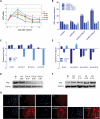
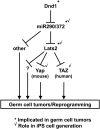
Pluripotent stem cells are derived from culture of early embryos or the germline and can be induced by reprogramming of somatic cells. Barriers to reprogramming that stabilize the differentiated state and have tumor suppression functions are expected to exist. However, we have a limited understanding of what such barriers might be. To find novel barriers to reprogramming to pluripotency, we compared the transcriptional profiles of the mouse germline with pluripotent and somatic cells, in vivo and in vitro. There is a remarkable global expression of the transcriptional program for pluripotency in primordial germ cells (PGCs). We identify parallels between PGC reprogramming to pluripotency and human germ cell tumorigenesis, including the loss of LATS2, a tumor suppressor kinase of the Hippo pathway. We show that knockdown of LATS2 increases the efficiency of induction of pluripotency in human cells. LATS2 RNAi, unlike p53 RNAi, specifically enhances the generation of fully reprogrammed iPS cells without accelerating cell proliferation. We further show that LATS2 represses reprogramming in human cells by post-transcriptionally antagonizing TAZ but not YAP, two downstream effectors of the Hippo pathway. These results reveal transcriptional parallels between germ cell transformation and the generation of iPS cells and indicate that the Hippo pathway constitutes a barrier to cellular reprogramming.
Figures
References
-
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. - PubMed
-
- Smith A.G. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. - PubMed
-
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

