Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development
- PMID: 22286183
- PMCID: PMC3320178
- DOI: 10.1104/pp.111.192104
Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development
Abstract
Several MADS box gene lineages involved in flower development have undergone duplications that correlate with the diversification of large groups of flowering plants. In the APETALA1 gene lineage, a major duplication coincides with the origin of the core eudicots, resulting in the euFUL and the euAP1 clades. Arabidopsis FRUITFULL (FUL) and APETALA1 (AP1) function redundantly in specifying floral meristem identity but function independently in sepal and petal identity (AP1) and in proper fruit development and determinacy (FUL). Many of these functions are largely conserved in other core eudicot euAP1 and euFUL genes, but notably, the role of APETALA1 as an "A-function" (sepal and petal identity) gene is thought to be Brassicaceae specific. Understanding how functional divergence of the core eudicot duplicates occurred requires a careful examination of the function of preduplication (FUL-like) genes. Using virus-induced gene silencing, we show that FUL-like genes in opium poppy (Papaver somniferum) and California poppy (Eschscholzia californica) function in axillary meristem growth and in floral meristem and sepal identity and that they also play a key role in fruit development. Interestingly, in opium poppy, these genes also control flowering time and petal identity, suggesting that AP1/FUL homologs might have been independently recruited in petal identity. Because the FUL-like gene functional repertoire encompasses all roles previously described for the core eudicot euAP1 and euFUL genes, we postulate subfunctionalization as the functional outcome after the major AP1/FUL gene lineage duplication event.
Figures


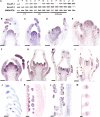


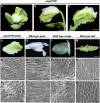
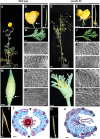
References
-
- Alvarez-Buylla ER, García-Ponce B, Garay-Arroyo A. (2006) Unique and redundant functional domains of APETALA1 and CAULIFLOWER, two recently duplicated Arabidopsis thaliana floral MADS-box genes. J Exp Bot 57: 3099–3107 - PubMed
-
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5: 569–579 - PubMed
-
- Becker A, Gleissberg S, Smyth DR. (2005) Floral and vegetative morphogenesis in California poppy (Eschscholzia californica Cham.). Int J Plant Sci 166: 537–555
-
- Becker A, Theissen G. (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

