Expression of Escherichia coli virulence usher protein attenuates wild-type Salmonella
- PMID: 22286706
- PMCID: PMC3337150
- DOI: 10.4161/viru.3.1.18447
Expression of Escherichia coli virulence usher protein attenuates wild-type Salmonella
Abstract
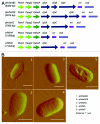
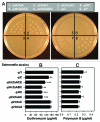
Generation of a live attenuated vaccine for bacterial pathogens often requires prior knowledge of the pathogen's virulence factors. We hypothesized an alternative approach of heterologous gene expression would make a wild-type (wt) pathogen more susceptible to host cell killing, thus, resulting in immunization. As proof of concept, the heterologous expression of enterotoxigenic E. coli (ETEC) colonization factor antigen I (CFA/I) was tested to attenuate Salmonella. The overexpression of CFA/I resulted in significant attenuation of wt Salmonella. In-depth studies revealed the attenuation depended on the co-expression of chaperone (CfaA) and usher (CfaC) proteins. Remarkably, the CfaAC-attenuated Salmonella conferred protection against wt Salmonella challenge. Mechanistic study indicated CfaAC made Salmonella outer membranes permeable, causing Salmonella to be vulnerable to host destruction. Thus, enhancing bacterial permeability via CfaAC represents an alternative method to attenuate pathogens despite the presence of unknown virulence factors.
Figures




Comment in
-
Coming of AGE: facile generation of attenuated vaccine strains through heterologous gene expression.Virulence. 2012 Jan-Feb;3(1):12-4. doi: 10.4161/viru.3.1.19087. Epub 2012 Jan 1. Virulence. 2012. PMID: 22286696 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
