A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver
- PMID: 22287103
- PMCID: PMC3317149
- DOI: 10.1101/gr.130286.111
A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver
Abstract
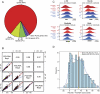
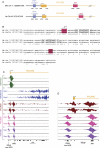
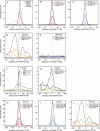
The genomic loci occupied by RNA polymerase (RNAP) III have been characterized in human culture cells by genome-wide chromatin immunoprecipitations, followed by deep sequencing (ChIP-seq). These studies have shown that only ∼40% of the annotated 622 human tRNA genes and pseudogenes are occupied by RNAP-III, and that these genes are often in open chromatin regions rich in active RNAP-II transcription units. We have used ChIP-seq to characterize RNAP-III-occupied loci in a differentiated tissue, the mouse liver. Our studies define the mouse liver RNAP-III-occupied loci including a conserved mammalian interspersed repeat (MIR) as a potential regulator of an RNAP-III subunit-encoding gene. They reveal that synteny relationships can be established between a number of human and mouse RNAP-III genes, and that the expression levels of these genes are significantly linked. They establish that variations within the A and B promoter boxes, as well as the strength of the terminator sequence, can strongly affect RNAP-III occupancy of tRNA genes. They reveal correlations with various genomic features that explain the observed variation of 81% of tRNA scores. In mouse liver, loci represented in the NCBI37/mm9 genome assembly that are clearly occupied by RNAP-III comprise 50 Rn5s (5S RNA) genes, 14 known non-tRNA RNAP-III genes, nine Rn4.5s (4.5S RNA) genes, and 29 SINEs. Moreover, out of the 433 annotated tRNA genes, half are occupied by RNAP-III. Transfer RNA gene expression levels reflect both an underlying genomic organization conserved in dividing human culture cells and resting mouse liver cells, and the particular promoter and terminator strengths of individual genes.
Figures


References
-
- Altman S, Wesolowski D, Puranam RS 1993. Nucleotide sequences of the RNA subunit of RNase P from several mammals. Genomics 18: 418–422 - PubMed
-
- Ame JC, Schreiber V, Fraulob V, Dolle P, de Murcia G, Niedergang CP 2001. A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. Structure and expression of the mouse PARP-2 gene. J Biol Chem 276: 11092–11099 - PubMed
-
- Anzai K, Kobayashi S, Kitamura N, Kanai Y, Nakajima H, Suehiro Y, Goto S 1986. Isolation and sequencing of a genomic clone for mouse brain specific small RNA. J Neurochem 47: 673–677 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
