A human XRCC4-XLF complex bridges DNA
- PMID: 22287571
- PMCID: PMC3287209
- DOI: 10.1093/nar/gks022
A human XRCC4-XLF complex bridges DNA
Abstract
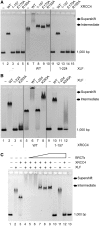
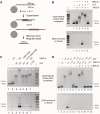
DNA double-strand breaks pose a significant threat to cell survival and must be repaired. In higher eukaryotes, such damage is repaired efficiently by non-homologous end joining (NHEJ). Within this pathway, XRCC4 and XLF fulfill key roles required for end joining. Using DNA-binding and -bridging assays, combined with direct visualization, we present evidence for how XRCC4-XLF complexes robustly bridge DNA molecules. This unanticipated, DNA Ligase IV-independent bridging activity by XRCC4-XLF suggests an early role for this complex during end joining, in addition to its more well-established later functions. Mutational analysis of the XRCC4-XLF C-terminal tail regions further identifies specialized functions in complex formation and interaction with DNA and DNA Ligase IV. Based on these data and the crystal structure of an extended protein filament of XRCC4-XLF at 3.94 Å, a model for XRCC4-XLF complex function in NHEJ is presented.
Figures




Comment on
- Nucleic Acids Res. doi: 10.1093/nar/gkr1315
References
-
- Roth DB, Nakagima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. - PubMed
-
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. - PubMed
-
- McElhinny SA, Havener JM, Garcia-Diaz M, Juárez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family × polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

