Skewed primary Igκ repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing
- PMID: 22287713
- PMCID: PMC3288532
- DOI: 10.4049/jimmunol.1103484
Skewed primary Igκ repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing
Abstract
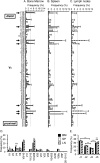
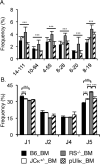
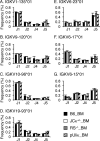
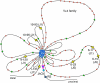
Previous estimates of the diversity of the mouse Ab repertoire have been based on fragmentary data as a result of many technical limitations, in particular, the many samples necessary to provide adequate coverage. In this study, we used 5'-coding end amplification of Igκ mRNAs from bone marrow, splenic, and lymph node B cells of C57BL/6 mice combined with amplicon pyrosequencing to assess the functional and nonfunctional Vκ repertoire. To evaluate the potential effects of receptor editing, we also compared V/J associations and usage in bone marrows of mouse mutants under constitutive negative selection or an altered ability to undergo secondary recombination. To focus on preimmune B cells, our cell sorting strategy excluded memory B cells and plasma cells. Analysis of ~90 Mbp, representing >250,000 individual transcripts from 59 mice, revealed that 101 distinct functional Vκ genes are used but at frequencies ranging from ~0.001 to ~10%. Usage of seven Vκ genes made up >40% of the repertoire. A small class of transcripts from apparently nonfunctional Vκ genes was found, as were occasional transcripts from several apparently functional genes that carry aberrant recombination signals. Of 404 potential V-J combinations (101 Vκs × 4 Jκs), 398 (98.5%) were found at least once in our sample. For most Vκ transcripts, all Jκs were used, but V-J association biases were common. Usage patterns were remarkably stable in different selective conditions. Overall, the primary κ repertoire is highly skewed by preferred rearrangements, limiting Ab diversity, but potentially facilitating receptor editing.
Figures




References
-
- Apel TW, Scherer A, Adachi T, Auch D, Ayane M, Reth M. The ribose 5-phosphate isomerase-encoding gene is located immediately downstream from that encoding murine immunoglobulin kappa. Gene. 1995;156:191–197. - PubMed
-
- Schable KF, Thiebe R, Bensch A, Brensing-Kuppers J, Heim V, Kirschbaum T, Lamm R, Ohnrich M, Pourrajabi S, Roschenthaler F, Schwendinger J, Wichelhaus D, Zocher I, Zachau HG. Characteristics of the immunoglobulin Vkappa genes, pseudogenes, relics and orphons in the mouse genome. Eur J Immunol. 1999;29:2082–2086. - PubMed
-
- Thiebe R, Schable KF, Bensch A, Brensing-Kuppers J, Heim V, Kirschbaum T, Mitlohner H, Ohnrich M, Pourrajabi S, Roschenthaler F, Schwendinger J, Wichelhaus D, Zocher I, Zachau HG. The variable genes and gene families of the mouse immunoglobulin kappa locus. Eur J Immunol. 1999;29:2072–2081. - PubMed
-
- Roschenthaler F, Kirschbaum T, Heim V, Kirschbaum V, Schable KF, Schwendinger J, Zocher I, Zachau HG. The 5' part of the mouse immunoglobulin kappa locus. Eur J Immunol. 1999;29:2065–2071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

