Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions
- PMID: 22289035
- PMCID: PMC3324619
- DOI: 10.1111/j.1600-0854.2012.01336.x
Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions
Abstract
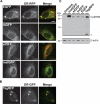
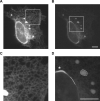
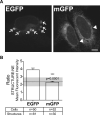
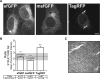
Several fluorescent proteins (FPs) are prone to forming low-affinity oligomers. This undesirable tendency is exacerbated when FPs are confined to membranes or when fused to naturally oligomeric proteins. Oligomerization of FPs limits their suitability for creating fusions with proteins of interest. Unfortunately, no standardized method evaluates the biologically relevant oligomeric state of FPs. Here, we describe a quantitative visual assay for assessing whether FPs are sufficiently monomeric under physiologic conditions. Membrane-associated FP-fusion proteins, by virtue of their constrained planar geometry, achieve high effective concentrations. We exploited this propensity to develop an assay to measure FP tendencies to oligomerize in cells. FPs were fused on the cytoplasmic end of an endoplasmic reticulum (ER) signal-anchor membrane protein (CytERM) and expressed in cells. Cells were scored based on the ability of CytERM to homo-oligomerize with proteins on apposing membranes and restructure the ER from a tubular network into organized smooth ER (OSER) whorl structures. The ratio of nuclear envelope and OSER structures mean fluorescent intensities for cells expressing enhanced green fluorescent protein (EGFP) or monomeric green fluorescent protein (mGFP) CytERM established standards for comparison of uncharacterized FPs. We tested three FPs and identified two as sufficiently monomeric, while a third previously reported as monomeric was found to strongly oligomerize.
© 2012 John Wiley & Sons A/S.
Figures

References
-
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. - PubMed
-
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–60. - PubMed
-
- Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

