Phosphorylation of BRN2 modulates its interaction with the Pax3 promoter to control melanocyte migration and proliferation
- PMID: 22290434
- PMCID: PMC3302439
- DOI: 10.1128/MCB.06257-11
Phosphorylation of BRN2 modulates its interaction with the Pax3 promoter to control melanocyte migration and proliferation
Abstract
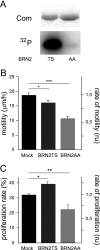
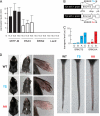
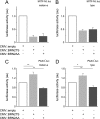
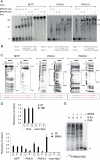
MITF-M and PAX3 are proteins central to the establishment and transformation of the melanocyte lineage. They control various cellular mechanisms, including migration and proliferation. BRN2 is a POU domain transcription factor expressed in melanoma cell lines and is involved in proliferation and invasion, at least in part by regulating the expression of MITF-M and PAX3. The T361 and S362 residues of BRN2, both in the POU domain, are conserved throughout the POU protein family and are targets for phosphorylation, but their roles in vivo remain unknown. To examine the role of this phosphorylation, we generated mutant BRN2 in which these two residues were replaced with alanines (BRN2TS→BRN2AA). When expressed in melanocytes in vitro or in the melanocyte lineage in transgenic mice, BRN2TS induced proliferation and repressed migration, whereas BRN2AA repressed both proliferation and migration. BRN2TS and BRN2AA bound and repressed the MITF-M promoter, whereas PAX3 transcription was induced by BRN2TS but repressed by BRN2AA. Expression of the BRN2AA transgene in a Mitf heterozygous background and in a Pax3 mutant background enhanced the coat color phenotype. Our findings show that melanocyte migration and proliferation are controlled both through the regulation of PAX3 by nonphosphorylated BRN2 and through the regulation of MITF-M by the overall BRN2 level.
Figures


References
-
- Abdel-Malek ZA, et al. 2001. The melanocortin 1 receptor is the principal mediator of the effects of agouti signaling protein on mammalian melanocytes. J. Cell Sci. 114:1019–1024 - PubMed
-
- Andersen B, Rosenfeld MG. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 22:2–35 - PubMed
-
- Arozarena I, et al. 2011. Oncogenic BRAF Induces Melanoma Cell Invasion by Downregulating the cGMP-Specific Phosphodiesterase PDE5A. Cancer Cell 19:45–57 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
