The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling
- PMID: 22290435
- PMCID: PMC3302445
- DOI: 10.1128/MCB.06222-11
The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling
Abstract
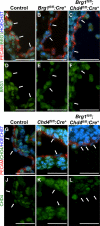
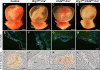
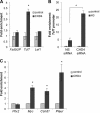
Canonical Wnt signaling plays an important role in embryonic and postnatal blood vessel development. We previously reported that the chromatin-remodeling enzyme BRG1 promotes vascular Wnt signaling. Vascular deletion of Brg1 results in aberrant yolk sac blood vessel morphology, which is rescued by pharmacological stimulation of Wnt signaling with lithium chloride (LiCl). We have now generated embryos lacking the chromatin-remodeling enzyme Chd4 in vascular endothelial cells. Unlike Brg1 mutants, Chd4 mutant embryos had normal yolk sac vascular morphology. However, concomitant deletion of Chd4 and Brg1 rescued vascular abnormalities seen in Brg1 mutant yolk sacs to the same extent as LiCl treatment. We hypothesized that Wnt signaling was upregulated in Chd4 mutant yolk sac vasculature. Indeed, we found that Chd4 deletion resulted in upregulation of the Wnt-responsive transcription factor Tcf7 and an increase in Wnt target gene expression in endothelial cells. Furthermore, we identified one Wnt target gene, Pitx2, that was downregulated in Brg1 mutant endothelial cells but was rescued following LiCl treatment and in Brg1 Chd4 double mutant vasculature, suggesting that PITX2 helps to mediate the restoration of yolk sac vascular remodeling under both conditions. We conclude that BRG1 and CHD4 antagonistically modulate Wnt signaling in developing yolk sac vessels to mediate normal vascular remodeling.
Figures




Similar articles
-
The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels.Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2282-7. doi: 10.1073/pnas.1013751108. Epub 2011 Jan 24. Proc Natl Acad Sci U S A. 2011. PMID: 21262838 Free PMC article.
-
Endothelial Chromatin-Remodeling Enzymes Regulate the Production of Critical ECM Components During Murine Lung Development.Arterioscler Thromb Vasc Biol. 2024 Aug;44(8):1784-1798. doi: 10.1161/ATVBAHA.124.320881. Epub 2024 Jun 13. Arterioscler Thromb Vasc Biol. 2024. PMID: 38868942 Free PMC article.
-
BRG1 (Brahma-Related Gene 1) Promotes Endothelial Mrtf Transcription to Establish Embryonic Capillary Integrity.Arterioscler Thromb Vasc Biol. 2017 Sep;37(9):1674-1682. doi: 10.1161/ATVBAHA.117.309785. Epub 2017 Jul 20. Arterioscler Thromb Vasc Biol. 2017. PMID: 28729363 Free PMC article.
-
Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer.Epigenetics. 2008 Mar-Apr;3(2):64-8. doi: 10.4161/epi.3.2.6153. Epub 2008 Apr 17. Epigenetics. 2008. PMID: 18437052 Review.
-
The Role of BRG1 in Antioxidant and Redox Signaling.Oxid Med Cell Longev. 2020 Sep 14;2020:6095673. doi: 10.1155/2020/6095673. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 33014273 Free PMC article. Review.
Cited by
-
The nucleosome remodeling and deacetylase-SWItch/sucrose non-fermentable antagonism regulates the coordinated activation of epithelial-to-mesenchymal transition and inflammation in oral cancer.J Natl Cancer Inst. 2025 Jul 1;117(7):1438-1455. doi: 10.1093/jnci/djaf065. J Natl Cancer Inst. 2025. PMID: 40112045 Free PMC article.
-
Human brain arteriovenous malformations express lymphatic-associated genes.Ann Clin Transl Neurol. 2014 Dec;1(12):982-95. doi: 10.1002/acn3.142. Epub 2014 Nov 18. Ann Clin Transl Neurol. 2014. PMID: 25574473 Free PMC article.
-
IL-1β Impacts Vascular Integrity and Lymphatic Function in the Embryonic Omentum.Circ Res. 2022 Feb 4;130(3):366-383. doi: 10.1161/CIRCRESAHA.121.319032. Epub 2022 Jan 6. Circ Res. 2022. PMID: 34986653 Free PMC article.
-
Integrated epigenomic analysis stratifies chromatin remodellers into distinct functional groups.Epigenetics Chromatin. 2019 Feb 12;12(1):12. doi: 10.1186/s13072-019-0258-9. Epigenetics Chromatin. 2019. PMID: 30755246 Free PMC article.
-
The NuRD chromatin-remodeling complex enzyme CHD4 prevents hypoxia-induced endothelial Ripk3 transcription and murine embryonic vascular rupture.Cell Death Differ. 2020 Feb;27(2):618-631. doi: 10.1038/s41418-019-0376-8. Epub 2019 Jun 24. Cell Death Differ. 2020. PMID: 31235857 Free PMC article.
References
-
- Bultman S, et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287–1295 - PubMed
-
- Chappell JC, Bautch VL. 2010. Vascular development: genetic mechanisms and links to vascular disease. Curr. Top. Dev. Biol. 90:43–72 - PubMed
-
- Dejana E. 2010. The role of wnt signaling in physiological and pathological angiogenesis. Circ. Res. 107:943–952 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
