The spt5 C-terminal region recruits yeast 3' RNA cleavage factor I
- PMID: 22290438
- PMCID: PMC3302448
- DOI: 10.1128/MCB.06310-11
The spt5 C-terminal region recruits yeast 3' RNA cleavage factor I
Abstract
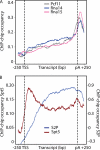
During transcription elongation, RNA polymerase II (Pol II) binds the general elongation factor Spt5. Spt5 contains a repetitive C-terminal region (CTR) that is required for cotranscriptional recruitment of the Paf1 complex (D. L. Lindstrom et al., Mol. Cell. Biol. 23:1368-1378, 2003; Z. Zhang, J. Fu, and D. S. Gilmour, Genes Dev. 19:1572-1580, 2005). Here we report a new role of the Spt5 CTR in the recruitment of 3' RNA-processing factors. Chromatin immunoprecipitation (ChIP) revealed that the Spt5 CTR is required for normal recruitment of pre-mRNA cleavage factor I (CFI) to the 3' ends of Saccharomyces cerevisiae genes. RNA contributes to CFI recruitment, as RNase treatment prior to ChIP further decreases CFI ChIP signals. Genome-wide ChIP profiling detected occupancy peaks of CFI subunits around 100 nucleotides downstream of the polyadenylation (pA) sites of genes. CFI recruitment to this defined region may result from simultaneous binding to the Spt5 CTR, to nascent RNA containing the pA sequence, and to the elongating Pol II isoform that is phosphorylated at serine 2 (S2) residues in its C-terminal domain (CTD). Consistent with this model, the CTR interacts with CFI in vitro but is not required for pA site recognition and transcription termination in vivo.
Figures



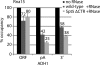


References
-
- Ahn SH, Kim M, Buratowski S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67–76 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
