Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan
- PMID: 22290972
- PMCID: PMC3318346
- DOI: 10.1128/AAC.05604-11
Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan
Abstract
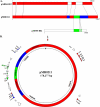
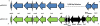
In response to global concerns over the spread of the New Delhi metallo-β-lactamase gene 1, bla(NDM-1), a monthly surveillance program was initiated in September 2010. All carbapenem-resistant Gram-negative strains forwarded to our facility are screened for this gene. To date, 321 carbapenem-resistant isolates, encompassing 11 bacterial species, have been tested. In February 2011, two strains of Providencia stuartii, submitted from a military hospital in Afghanistan, tested positive for bla(NDM-1). Both strains were identical by pulsed-field gel electrophoresis (PFGE). bla(NDM-1) was carried on a large plasmid, pMR0211, which was sequenced by emulsion PCR and pyrosequencing. pMR0211 is 178,277 bp in size and belongs to incompatibility group A/C. The plasmid consists of a backbone with considerable homology to pAR060302 from Escherichia coli, and it retains many of the antibiotic resistance genes associated with it. The plasmid also shares common elements with the pNDM-HK plasmid, including bla(NDM-1), armA, and sul1. However, gene orientation is reversed, and a 3-kb fragment from this region is absent from pMR0211. pMR0211 also contains additional genes, including the aminoglycoside-modifying enzyme loci aadA and aac(6'), the quinolone resistance gene qnrA, a gene with highest homology to a U32 family peptidase from Shewanella amazonensis, and the bla(OXA-10) gene. The finding of this gene in an intrinsically colistin-resistant species such as Providencia stuartii is especially worrisome, as it renders the organism resistant to nearly every available antibiotic. The presence of multiple insertion sequences and transposons flanking the region containing the bla(NDM-1) gene further highlights the potential mobility associated with this gene.
Figures
References
-
- Bonomo RA. 2011. New Delhi metallo-beta-lactamase and multidrug resistance: a global SOS? Clin. Infect. Dis. 52:485–487 - PubMed
-
- Boucher HW, et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 - PubMed
-
- Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 - PubMed
-
- Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

