A 5'-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli
- PMID: 22291205
- PMCID: PMC3285938
- DOI: 10.1261/rna.027698.111
A 5'-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli
Abstract
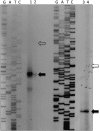
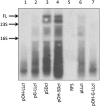
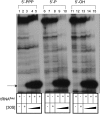
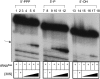
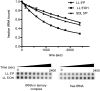
The bacteriophage λ's cI mRNA was utilized to examine the importance of the 5'-terminal phosphate on expression of leadered and leaderless mRNA in Escherichia coli. A hammerhead ribozyme was used to produce leadered and leaderless mRNAs, in vivo and in vitro, that contain a 5'-hydroxyl. Although these mRNAs may not occur naturally in the bacterial cell, they allow for the study of the importance of the 5'-phosphorylation state in ribosome binding and translation of leadered and leaderless mRNAs. Analyses with mRNAs containing either a 5'-phosphate or a 5'-hydroxyl indicate that leaderless cI mRNA requires a 5'-phosphate for stable ribosome binding in vitro as well as expression in vivo. Ribosome-binding assays show that 30S subunits and 70S ribosomes do not bind as strongly to 5'-hydroxyl as they do to 5'-phosphate containing leaderless mRNA and the tRNA-dependent ternary complex is less stable. Additionally, filter-binding assays revealed that the 70S ternary complex formed with a leaderless mRNA containing a 5'-hydroxyl has a dissociation rate (k(off)) that is 4.5-fold higher compared with the complex formed with a 5'-phosphate leaderless mRNA. Fusion to a lacZ reporter gene revealed that leaderless cI mRNA expression with a 5'-hydroxyl was >100-fold lower than the equivalent mRNA with a 5'-phosphate. These data indicate that a 5'-phosphate is an important feature of leaderless mRNA for stable ribosome binding and expression.
Figures



Similar articles
-
Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5'-terminal AUG.RNA. 2008 Oct;14(10):2159-69. doi: 10.1261/rna.1089208. Epub 2008 Aug 28. RNA. 2008. PMID: 18755843 Free PMC article.
-
Temperature-dependent translation of leaderless and canonical mRNAs in Escherichia coli.FEMS Microbiol Lett. 2002 Jun 4;211(2):161-7. doi: 10.1111/j.1574-6968.2002.tb11219.x. FEMS Microbiol Lett. 2002. PMID: 12076807
-
Isolation and characterization of ribosomes and translation initiation factors from the gram-positive soil bacterium Streptomyces lividans.J Bacteriol. 2004 Oct;186(20):6864-75. doi: 10.1128/JB.186.20.6864-6875.2004. J Bacteriol. 2004. PMID: 15466040 Free PMC article.
-
Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control.Mol Microbiol. 2002 Jan;43(1):239-46. doi: 10.1046/j.1365-2958.2002.02739.x. Mol Microbiol. 2002. PMID: 11849551 Review.
-
Translation initiation and the fate of bacterial mRNAs.FEMS Microbiol Rev. 2006 Nov;30(6):967-79. doi: 10.1111/j.1574-6976.2006.00043.x. Epub 2006 Sep 21. FEMS Microbiol Rev. 2006. PMID: 16989654 Review.
Cited by
-
Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.Nat Commun. 2025 Jan 2;16(1):348. doi: 10.1038/s41467-024-55718-5. Nat Commun. 2025. PMID: 39753558 Free PMC article.
-
Leaderless mRNAs in the Spotlight: Ancient but Not Outdated!Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0016-2017. doi: 10.1128/microbiolspec.RWR-0016-2017. Microbiol Spectr. 2018. PMID: 30006995 Free PMC article. Review.
-
Translational regulation in mycobacteria and its implications for pathogenicity.Nucleic Acids Res. 2018 Aug 21;46(14):6950-6961. doi: 10.1093/nar/gky574. Nucleic Acids Res. 2018. PMID: 29947784 Free PMC article. Review.
-
How Dedicated Ribosomes Translate a Leaderless mRNA.J Mol Biol. 2024 Feb 15;436(4):168423. doi: 10.1016/j.jmb.2023.168423. Epub 2024 Jan 5. J Mol Biol. 2024. PMID: 38185325 Free PMC article.
-
Noncanonical features and modifications on the 5'-end of bacterial sRNAs and mRNAs.Wiley Interdiscip Rev RNA. 2019 Mar;10(2):e1509. doi: 10.1002/wrna.1509. Epub 2018 Oct 1. Wiley Interdiscip Rev RNA. 2019. PMID: 30276982 Free PMC article. Review.
References
-
- Chang B, Halgamuge S, Tang S 2006. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene 373: 90–99 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
