Reduced lentivirus susceptibility in sheep with TMEM154 mutations
- PMID: 22291605
- PMCID: PMC3266874
- DOI: 10.1371/journal.pgen.1002467
Reduced lentivirus susceptibility in sheep with TMEM154 mutations
Abstract
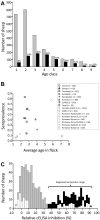
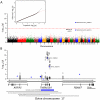
Visna/Maedi, or ovine progressive pneumonia (OPP) as it is known in the United States, is an incurable slow-acting disease of sheep caused by persistent lentivirus infection. This disease affects multiple tissues, including those of the respiratory and central nervous systems. Our aim was to identify ovine genetic risk factors for lentivirus infection. Sixty-nine matched pairs of infected cases and uninfected controls were identified among 736 naturally exposed sheep older than five years of age. These pairs were used in a genome-wide association study with 50,614 markers. A single SNP was identified in the ovine transmembrane protein (TMEM154) that exceeded genome-wide significance (unadjusted p-value 3×10(-9)). Sanger sequencing of the ovine TMEM154 coding region identified six missense and two frameshift deletion mutations in the predicted signal peptide and extracellular domain. Two TMEM154 haplotypes encoding glutamate (E) at position 35 were associated with infection while a third haplotype with lysine (K) at position 35 was not. Haplotypes encoding full-length E35 isoforms were analyzed together as genetic risk factors in a multi-breed, matched case-control design, with 61 pairs of 4-year-old ewes. The odds of infection for ewes with one copy of a full-length TMEM154 E35 allele were 28 times greater than the odds for those without (p-value<0.0001, 95% CI 5-1,100). In a combined analysis of nine cohorts with 2,705 sheep from Nebraska, Idaho, and Iowa, the relative risk of infection was 2.85 times greater for sheep with a full-length TMEM154 E35 allele (p-value<0.0001, 95% CI 2.36-3.43). Although rare, some sheep were homozygous for TMEM154 deletion mutations and remained uninfected despite a lifetime of significant exposure. Together, these findings indicate that TMEM154 may play a central role in ovine lentivirus infection and removing sheep with the most susceptible genotypes may help eradicate OPP and protect flocks from reinfection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Büchen-Osmond C, editor. ICTVdB Management 00.061.1.06.008. Visna/maedi virus. New York: Columbia University; 2006.
-
- Petursson G. Experience with visna virus in Iceland. Ann N Y Acad Sci. 1994;724:43–49. - PubMed
-
- Greenwood PL, North RN, Kirkland PD. Prevalence, spread and control of caprine arthritis-encephalitis virus in dairy goat herds in New South Wales. Aust Vet J. 1995;72:341–345. - PubMed
-
- Motha MX, Ralston JC. Evaluation of ELISA for detection of antibodies to CAEV in milk. Vet Microbiol. 1994;38:359–367. - PubMed
-
- USDAAnimal and Plant Health Inspection Service VS, Centers for Epidemiology and Animal Health, editor. Fort Collins; 2003. Ovine Progressive Pneumonia: Awareness, Management, and Seroprevalence.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

