Sex pheromone evolution is associated with differential regulation of the same desaturase gene in two genera of leafroller moths
- PMID: 22291612
- PMCID: PMC3266893
- DOI: 10.1371/journal.pgen.1002489
Sex pheromone evolution is associated with differential regulation of the same desaturase gene in two genera of leafroller moths
Abstract
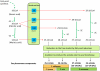
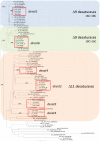
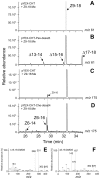
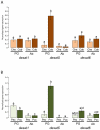
Chemical signals are prevalent in sexual communication systems. Mate recognition has been extensively studied within the Lepidoptera, where the production and recognition of species-specific sex pheromone signals are typically the defining character. While the specific blend of compounds that makes up the sex pheromones of many species has been characterized, the molecular mechanisms underpinning the evolution of pheromone-based mate recognition systems remain largely unknown. We have focused on two sets of sibling species within the leafroller moth genera Ctenopseustis and Planotortrix that have rapidly evolved the use of distinct sex pheromone blends. The compounds within these blends differ almost exclusively in the relative position of double bonds that are introduced by desaturase enzymes. Of the six desaturase orthologs isolated from all four species, functional analyses in yeast and gene expression in pheromone glands implicate three in pheromone biosynthesis, two Δ9-desaturases, and a Δ10-desaturase, while the remaining three desaturases include a Δ6-desaturase, a terminal desaturase, and a non-functional desaturase. Comparative quantitative real-time PCR reveals that the Δ10-desaturase is differentially expressed in the pheromone glands of the two sets of sibling species, consistent with differences in the pheromone blend in both species pairs. In the pheromone glands of species that utilize (Z)-8-tetradecenyl acetate as sex pheromone component (Ctenopseustis obliquana and Planotortrix octo), the expression levels of the Δ10-desaturase are significantly higher than in the pheromone glands of their respective sibling species (C. herana and P. excessana). Our results demonstrate that interspecific sex pheromone differences are associated with differential regulation of the same desaturase gene in two genera of moths. We suggest that differential gene regulation among members of a multigene family may be an important mechanism of molecular innovation in sex pheromone evolution and speciation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. - PubMed
-
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. - DOI - PMC - PubMed
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. - PubMed
-
- King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed

