TRIM5α and Species Tropism of HIV/SIV
- PMID: 22291694
- PMCID: PMC3264904
- DOI: 10.3389/fmicb.2012.00013
TRIM5α and Species Tropism of HIV/SIV
Abstract
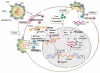
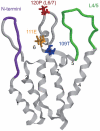
Human immunodeficiency virus type 1 (HIV-1) infects humans and chimpanzees but not old world monkeys (OWMs) such as the rhesus monkey (Rh) and cynomolgus monkey (CM). HIV-1 efficiently enters cells of OWMs but encounters a block before reverse transcription. This narrow host range is attributed to a barrier in the host cell. In 2004, the screening of a Rh cDNA library identified tripartite motif 5α (TRIM5α) as a cellular antiviral factor. TRIM5α is one of splicing variants produced by TRIM5 gene and TRIM5 proteins are members of the TRIM family containing RING, B-box 2, and coiled-coil domains. The RING domain is frequently found in E3 ubiquitin ligase and TRIM5α is degraded via the ubiquitin-proteasome-dependent pathway. Among TRIM5 splicing variants, TRIM5α alone has an additional C-terminal PRYSPRY (B30.2) domain. Previous studies have shown that sequence variation in variable regions of the PRYSPRY domain among different monkey species affects species-specific retrovirus infection, while amino acid sequence differences in the viral capsid protein determine viral sensitivity to restriction. TRIM5α recognizes the multimerized capsid proteins (viral core) of an incoming virus by its PRYSPRY domain and is thus believed to control retroviral infection. There are significant intraspecies variations in the Rh-TRIM5 gene. It has also been reported that some Rh and CM individuals have retrotransposed cyclophilin A open reading frame in the TRIM5 gene, which produces TRIM5-cyclophilin A fusion protein (TRIMCyp). TRIMCyp, which was originally identified as an anti-HIV-1 factor of New World owl monkeys, is an interesting example of the gain of a new function by retrotransposition. As different TRIM5 genotypes of Rh showed different levels of simian immunodeficiency virus replication in vivo, the TRIM5 genotyping is thought to be important in acquired immunodeficiency syndrome monkey models.
Keywords: HIV-1; HIV-2; SIV; TRIM5α; TRIMCyp; cynomolgus monkey; rhesus monkey.
Figures



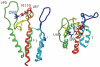
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

