Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium
- PMID: 22291916
- PMCID: PMC3266884
- DOI: 10.1371/journal.pone.0030187
Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium
Abstract
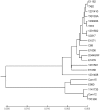
Recent studies have pointed to the existence of two subpopulations of Enterococcus faecium, one containing primarily commensal/community-associated (CA) strains and one that contains most clinical or hospital-associated (HA) strains, including those classified by multi-locus sequence typing (MLST) as belonging to the CC17 group. The HA subpopulation more frequently has IS16, pathogenicity island(s), and plasmids or genes associated with antibiotic resistance, colonization, and/or virulence. Supporting the two clades concept, we previously found a 3-10% difference between four genes from HA-clade strains vs. CA-clade strains, including 5% difference between pbp5-R of ampicillin-resistant, HA strains and pbp5-S of ampicillin-sensitive, CA strains. To further investigate the core genome of these subpopulations, we studied 100 genes from 21 E. faecium genome sequences; our analyses of concatenated sequences, SNPs, and individual genes all identified two distinct groups. With the concatenated sequence, HA-clade strains differed by 0-1% from one another while CA clade strains differed from each other by 0-1.1%, with 3.5-4.2% difference between the two clades. While many strains had a few genes that grouped in one clade with most of their genes in the other clade, one strain had 28% of its genes in the CA clade and 72% in the HA clade, consistent with the predicted role of recombination in the evolution of E. faecium. Using estimates for Escherichia coli, molecular clock calculations using sSNP analysis indicate that these two clades may have diverged ≥1 million years ago or, using the higher mutation rate for Bacillus anthracis, ∼300,000 years ago. These data confirm the existence of two clades of E. faecium and show that the differences between the HA and CA clades occur at the core genomic level and long preceded the modern antibiotic era.
Conflict of interest statement
Figures

References
-
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. - PubMed
-
- Klare I, Konstabel C, Badstubner D, Werner G, Witte W. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int J Food Microbiol. 2003;88:269–290. - PubMed
-
- Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006;9:454–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

