The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes
- PMID: 22291979
- PMCID: PMC3264610
- DOI: 10.1371/journal.pone.0030531
The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes
Abstract
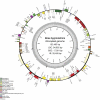
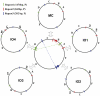
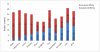
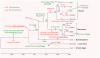
The complete nucleotide sequences of the chloroplast (cp) and mitochondrial (mt) genomes of resurrection plant Boea hygrometrica (Bh, Gesneriaceae) have been determined with the lengths of 153,493 bp and 510,519 bp, respectively. The smaller chloroplast genome contains more genes (147) with a 72% coding sequence, and the larger mitochondrial genome have less genes (65) with a coding faction of 12%. Similar to other seed plants, the Bh cp genome has a typical quadripartite organization with a conserved gene in each region. The Bh mt genome has three recombinant sequence repeats of 222 bp, 843 bp, and 1474 bp in length, which divide the genome into a single master circle (MC) and four isomeric molecules. Compared to other angiosperms, one remarkable feature of the Bh mt genome is the frequent transfer of genetic material from the cp genome during recent Bh evolution. We also analyzed organellar genome evolution in general regarding genome features as well as compositional dynamics of sequence and gene structure/organization, providing clues for the understanding of the evolution of organellar genomes in plants. The cp-derived sequences including tRNAs found in angiosperm mt genomes support the conclusion that frequent gene transfer events may have begun early in the land plant lineage.
Conflict of interest statement
Figures

References
-
- Olmstead R, Palmer J. Chloroplast DNA systematics: A review of methods and data analysis. American journal of botany. 1994;81:1205–1224.
-
- Qiu Y-L, Li L, Wang B, Xue J-Y, Hendry TA, et al. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. Journal of Systematics and Evolution. 2010;48:391–425.
-
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution. 2006;23:2175–2190. - PubMed
-
- Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, et al. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 2007;45:547–563. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

