Nurr1 protein is required for N-methyl-D-aspartic acid (NMDA) receptor-mediated neuronal survival
- PMID: 22294685
- PMCID: PMC3322860
- DOI: 10.1074/jbc.M111.272427
Nurr1 protein is required for N-methyl-D-aspartic acid (NMDA) receptor-mediated neuronal survival
Abstract
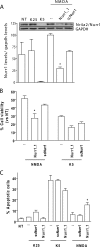
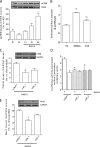
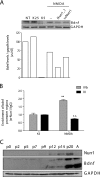
NMDA receptor (NMDAR) stimulation promotes neuronal survival during brain development. Cerebellar granule cells (CGCs) need NMDAR stimulation to survive and develop. These neurons differentiate and mature during its migration from the external granular layer to the internal granular layer, and lack of excitatory inputs triggers their apoptotic death. It is possible to mimic this process in vitro by culturing CGCs in low KCl concentrations (5 mm) in the presence or absence of NMDA. Using this experimental approach, we have obtained whole genome expression profiles after 3 and 8 h of NMDA addition to identify genes involved in NMDA-mediated survival of CGCs. One of the identified genes was Nurr1, a member of the orphan nuclear receptor subfamily Nr4a. Our results report a direct regulation of Nurr1 by CREB after NMDAR stimulation. ChIP assay confirmed CREB binding to Nurr1 promoter, whereas CREB shRNA blocked NMDA-mediated increase in Nurr1 expression. Moreover, we show that Nurr1 is important for NMDAR survival effect. We show that Nurr1 binds to Bdnf promoter IV and that silencing Nurr1 by shRNA leads to a decrease in brain-derived neurotrophic factor (BDNF) protein levels and a reduction of NMDA neuroprotective effect. Also, we report that Nurr1 and BDNF show a similar expression pattern during postnatal cerebellar development. Thus, we conclude that Nurr1 is a downstream target of CREB and that it is responsible for the NMDA-mediated increase in BDNF, which is necessary for the NMDA-mediated prosurvival effect on neurons.
Figures

References
-
- Gould E., Cameron H. A., McEwen B. S. (1994) Blockade of NMDA receptors increases cell death and birth in the developing rat dentate gyrus. J. Comp. Neurol. 340, 551–565 - PubMed
-
- Ikonomidou C., Bosch F., Miksa M., Bittigau P., Vöckler J., Dikranian K., Tenkova T. I., Stefovska V., Turski L., Olney J. W. (1999) Blockade of NMDA receptors and apoptotic neurodegeneration in the developing brain. Science 283, 70–74 - PubMed
-
- Balázs R., Jørgensen O. S., Hack N. (1988) N-Methyl-d-aspartate promotes the survival of cerebellar granule cells in culture. Neuroscience 27, 437–451 - PubMed
-
- Altman J. (1972) Postnatal development of the cerebellar cortex in the rat. 3: maturation of the components of the granular layer. J. Comp. Neurol. 145, 465–513 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

