Genome-wide repression of NF-κB target genes by transcription factor MIBP1 and its modulation by O-linked β-N-acetylglucosamine (O-GlcNAc) transferase
- PMID: 22294689
- PMCID: PMC3323019
- DOI: 10.1074/jbc.M111.298521
Genome-wide repression of NF-κB target genes by transcription factor MIBP1 and its modulation by O-linked β-N-acetylglucosamine (O-GlcNAc) transferase
Abstract
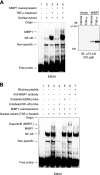
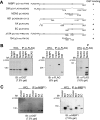
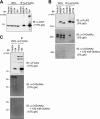
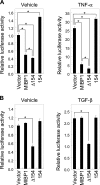
The transcription factor c-MYC intron binding protein 1 (MIBP1) binds to various genomic regulatory regions, including intron 1 of c-MYC. This factor is highly expressed in postmitotic neurons in the fetal brain and may be involved in various biological steps, such as neurological and immunological processes. In this study, we globally characterized the transcriptional targets of MIBP1 and proteins that interact with MIBP1. Microarray hybridization followed by gene set enrichment analysis revealed that genes involved in the pathways downstream of MYC, NF-κB, and TGF-β were down-regulated when HEK293 cells stably overexpressed MIBP1. In silico transcription factor binding site analysis of the promoter regions of these down-regulated genes showed that the NF-κB binding site was the most overrepresented. The up-regulation of genes known to be in the NF-κB pathway after the knockdown of endogenous MIBP1 in HT1080 cells supports the view that MIBP1 is a down-regulator of the NF-κB pathway. We also confirmed the binding of the MIBP1 to the NF-κB site. By immunoprecipitation and mass spectrometry, we detected O-linked β-N-acetylglucosamine (O-GlcNAc) transferase as a prominent binding partner of MIBP1. Analyses using deletion mutants revealed that a 154-amino acid region of MIBP1 was necessary for its O-GlcNAc transferase binding and O-GlcNAcylation. A luciferase reporter assay showed that NF-κB-responsive expression was repressed by MIBP1, and stronger repression by MIBP1 lacking the 154-amino acid region was observed. Our results indicate that the primary effect of MIBP1 expression is the down-regulation of the NF-κB pathway and that this effect is attenuated by O-GlcNAc signaling.
Figures

Similar articles
-
Characterization of the biological functions of a transcription factor, c-myc intron binding protein 1 (MIBP1).J Biochem. 2002 Mar;131(3):349-57. doi: 10.1093/oxfordjournals.jbchem.a003109. J Biochem. 2002. PMID: 11872163
-
Glucosamine improves cardiac function following trauma-hemorrhage by increased protein O-GlcNAcylation and attenuation of NF-{kappa}B signaling.Am J Physiol Heart Circ Physiol. 2009 Feb;296(2):H515-23. doi: 10.1152/ajpheart.01025.2008. Epub 2008 Dec 19. Am J Physiol Heart Circ Physiol. 2009. PMID: 19098112 Free PMC article.
-
Triptolide-induced cell death in pancreatic cancer is mediated by O-GlcNAc modification of transcription factor Sp1.J Biol Chem. 2013 Nov 22;288(47):33927-33938. doi: 10.1074/jbc.M113.500983. Epub 2013 Oct 15. J Biol Chem. 2013. PMID: 24129563 Free PMC article.
-
Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity.Am J Physiol Endocrinol Metab. 2008 Jul;295(1):E17-28. doi: 10.1152/ajpendo.90281.2008. Epub 2008 Apr 29. Am J Physiol Endocrinol Metab. 2008. PMID: 18445751 Free PMC article. Review.
-
Regulation of cancer metabolism by O-GlcNAcylation.Glycoconj J. 2014 Apr;31(3):185-91. doi: 10.1007/s10719-013-9515-5. Epub 2013 Dec 10. Glycoconj J. 2014. PMID: 24323367 Review.
Cited by
-
Hyperglycemia and aberrant O-GlcNAcylation: contributions to tumor progression.J Bioenerg Biomembr. 2018 Jun;50(3):175-187. doi: 10.1007/s10863-017-9740-x. Epub 2018 Jan 11. J Bioenerg Biomembr. 2018. PMID: 29322286 Review.
-
O-linked β-N-acetylglucosamine (O-GlcNAc) modification: Emerging pathogenesis and a therapeutic target of diabetic nephropathy.Diabet Med. 2025 Feb;42(2):e15436. doi: 10.1111/dme.15436. Epub 2024 Sep 16. Diabet Med. 2025. PMID: 39279604 Free PMC article. Review.
-
Loss-of-function variants in HIVEP2 are a cause of intellectual disability.Eur J Hum Genet. 2016 Apr;24(4):556-61. doi: 10.1038/ejhg.2015.151. Epub 2015 Jul 8. Eur J Hum Genet. 2016. PMID: 26153216 Free PMC article.
-
CircHivep2 contributes to microglia activation and inflammation via miR-181a-5p/SOCS2 signalling in mice with kainic acid-induced epileptic seizures.J Cell Mol Med. 2020 Nov;24(22):12980-12993. doi: 10.1111/jcmm.15894. Epub 2020 Oct 1. J Cell Mol Med. 2020. PMID: 33002329 Free PMC article.
-
Identification and characterization of human MIBP1 gene in glioma cell differentiation.J Mol Neurosci. 2014 Feb;52(2):294-301. doi: 10.1007/s12031-013-0144-z. Epub 2013 Oct 26. J Mol Neurosci. 2014. PMID: 24158731
References
-
- Vaquerizas J. M., Kummerfeld S. K., Teichmann S. A., Luscombe N. M. (2009) A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10, 252–263 - PubMed
-
- Singh H., LeBowitz J. H., Baldwin A. S., Jr., Sharp P. A. (1988) Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell 52, 415–423 - PubMed
-
- Fan C. M., Maniatis T. (1990) A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes Dev. 4, 29–42 - PubMed
-
- Allen C. E., Wu L. C. (2005) in Zinc Finger Proteins (Iuchi S., Kuldell N., eds) pp. 213–220, Springer, New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

