Personalized cardiovascular medicine and drug development: time for a new paradigm
- PMID: 22294708
- PMCID: PMC3273851
- DOI: 10.1161/CIRCULATIONAHA.111.089243
Personalized cardiovascular medicine and drug development: time for a new paradigm
Figures
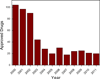




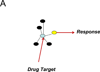



References
-
- Weiss S. Routine Practices: Medical Service of the Peter Bent Brigham Hospital. Self-published; Boston: 1940. pp. 143–145.
-
- Ridley M. Drugs That Are as Smart as Our Diseases. The Wall Street Journal. 2011 September 17;
-
- Pammolli F, Magazzini L, Riccaboni M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Disc. 2011;10:428–438. - PubMed
-
- Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Disc. 2011;10:507–519. - PubMed
-
- Loscalzo J. Homocysteine trials—clear outcomes for complex reasons. N. Engl. J. Med. 2006;354:1629–1632. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

