Nlrp2, a maternal effect gene required for early embryonic development in the mouse
- PMID: 22295082
- PMCID: PMC3266252
- DOI: 10.1371/journal.pone.0030344
Nlrp2, a maternal effect gene required for early embryonic development in the mouse
Abstract
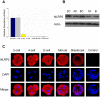
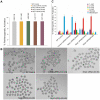
Maternal effect genes encode proteins that are produced during oogenesis and play an essential role during early embryogenesis. Genetic ablation of such genes in oocytes can result in female subfertility or infertility. Here we report a newly identified maternal effect gene, Nlrp2, which plays a role in early embryogenesis in the mouse. Nlrp2 mRNAs and their proteins (∼118 KDa) are expressed in oocytes and granulosa cells during folliculogenesis. The transcripts show a striking decline in early preimplantation embryos before zygotic genome activation, but the proteins remain present through to the blastocyst stage. Immunogold electron microscopy revealed that the NLRP2 protein is located in the cytoplasm, nucleus and close to nuclear pores in the oocytes, as well as in the surrounding granulosa cells. Using RNA interference, we knocked down Nlrp2 transcription specifically in mouse germinal vesicle oocytes. The knockdown oocytes could progress through the metaphase of meiosis I and emit the first polar body. However, the development of parthenogenetic embryos derived from Nlrp2 knockdown oocytes mainly blocked at the 2-cell stage. The maternal depletion of Nlrp2 in zygotes led to early embryonic arrest. In addition, overexpression of Nlrp2 in zygotes appears to lead to normal development, but increases blastomere apoptosis in blastocysts. These results provide the first evidence that Nlrp2 is a member of the mammalian maternal effect genes and required for early embryonic development in the mouse.
Conflict of interest statement
Figures



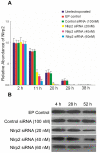

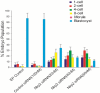
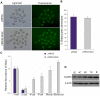
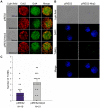
References
-
- Latham KE. Mechanisms and control of embryonic genome activation in mammalian embryos. Int Rev Cytol. 1999;193:71–124. - PubMed
-
- Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407:693–694. - PubMed
-
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26:267–268. - PubMed
-
- Nusslein-Volhard C, Lohs-Schardin M, Sander K, Cremer C. A dorso-ventral shift of embryonic primordia in a new maternal-effect mutant of Drosophila. Nature. 1980;283:474–476. - PubMed
-
- Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

