Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease
- PMID: 22295904
- PMCID: PMC3391577
- DOI: 10.1021/ja2095766
Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease
Abstract
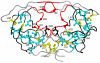
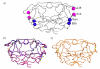
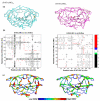
Human immunodeficiency virus Type-1 (HIV-1) protease is crucial for viral maturation and infectivity. Studies of protease dynamics suggest that the rearrangement of the hydrophobic core is essential for enzyme activity. Many mutations in the hydrophobic core are also associated with drug resistance and may modulate the core flexibility. To test the role of flexibility in protease activity, pairs of cysteines were introduced at the interfaces of flexible regions remote from the active site. Disulfide bond formation was confirmed by crystal structures and by alkylation of free cysteines and mass spectrometry. Oxidized and reduced crystal structures of these variants show the overall structure of the protease is retained. However, cross-linking the cysteines led to drastic loss in enzyme activity, which was regained upon reducing the disulfide cross-links. Molecular dynamics simulations showed that altered dynamics propagated throughout the enzyme from the engineered disulfide. Thus, altered flexibility within the hydrophobic core can modulate HIV-1 protease activity, supporting the hypothesis that drug resistant mutations distal from the active site can alter the balance between substrate turnover and inhibitor binding by modulating enzyme activity.
© 2012 American Chemical Society
Figures

References
-
- Kramer RA, Schaber MD, Skalka AM, Ganguly K, Wong-Staal F, Reddy EP. Science. 1986;231:1580–4. - PubMed
-
- Muzammil S, Ross P, Freire E. Biochemistry. 2003;42:631–8. - PubMed
-
- Ohtaka H, Schon A, Freire E. Biochemistry. 2003;42:13659–66. - PubMed
-
- Clemente JC, Moose RE, Hemrajani R, Whitford LR, Govindasamy L, Reutzel R, McKenna R, Agbandje-McKenna M, Goodenow MM, Dunn BM. Biochemistry. 2004;43:12141–12151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

