Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity
- PMID: 22296846
- PMCID: PMC3283125
- DOI: 10.1242/dev.077362
Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity
Abstract
A subfamily of Drosophila homeodomain (HD) transcription factors (TFs) controls the identities of individual muscle founder cells (FCs). However, the molecular mechanisms by which these TFs generate unique FC genetic programs remain unknown. To investigate this problem, we first applied genome-wide mRNA expression profiling to identify genes that are activated or repressed by the muscle HD TFs Slouch (Slou) and Muscle segment homeobox (Msh). Next, we used protein-binding microarrays to define the sequences that are bound by Slou, Msh and other HD TFs that have mesodermal expression. These studies revealed that a large class of HDs, including Slou and Msh, predominantly recognize TAAT core sequences but that each HD also binds to unique sites that deviate from this canonical motif. To understand better the regulatory specificity of an individual FC identity HD, we evaluated the functions of atypical binding sites that are preferentially bound by Slou relative to other HDs within muscle enhancers that are either activated or repressed by this TF. These studies showed that Slou regulates the activities of particular myoblast enhancers through Slou-preferred sequences, whereas swapping these sequences for sites that are capable of binding to multiple HD family members does not support the normal regulatory functions of Slou. Moreover, atypical Slou-binding sites are overrepresented in putative enhancers associated with additional Slou-responsive FC genes. Collectively, these studies provide new insights into the roles of individual HD TFs in determining cellular identity, and suggest that the diversity of HD binding preferences can confer regulatory specificity.
Figures
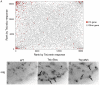

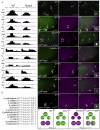
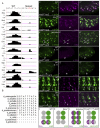
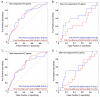
References
-
- Azpiazu N., Frasch M. (1993). tinman and bagpipe - two homeobox genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325–1340 - PubMed
-
- Barolo S., Carver L. A., Posakony J. W. (2000). GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726–732 - PubMed
-
- Baylies M. K., Arias A. M., Bate M. (1995). wingless is required for the formation of a subset of muscle founder cells during Drosophila embryogenesis. Development 121, 3829–3837 - PubMed
-
- Baylies M. K., Bate M., Ruiz Gomez M. (1998). Myogenesis: a view from Drosophila. Cell 93, 921–927 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

