Autophagic degradation of farnesylated prelamin A as a therapeutic approach to lamin-linked progeria
- PMID: 22297442
- PMCID: PMC3284238
- DOI: 10.4081/ejh.2011.e36
Autophagic degradation of farnesylated prelamin A as a therapeutic approach to lamin-linked progeria
Erratum in
- Eur J Histochem. 2013;57(4):e42
Abstract
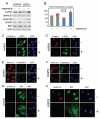
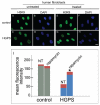
Farnesylated prelamin A is a processing intermediate produced in the lamin A maturation pathway. Accumulation of a truncated farnesylated prelamin A form, called progerin, is a hallmark of the severe premature ageing syndrome, Hutchinson-Gilford progeria. Progerin elicits toxic effects in cells, leading to chromatin damage and cellular senescence and ultimately causes skin and endothelial defects, bone resorption, lipodystrophy and accelerated ageing. Knowledge of the mechanism underlying prelamin A turnover is critical for the development of clinically effective protein inhibitors that can avoid accumulation to toxic levels without impairing lamin A/C expression, which is essential for normal biological functions. Little is known about specific molecules that may target farnesylated prelamin A to elicit protein degradation. Here, we report the discovery of rapamycin as a novel inhibitor of progerin, which dramatically and selectively decreases protein levels through a mechanism involving autophagic degradation. Rapamycin treatment of progeria cells lowers progerin, as well as wild-type prelamin A levels, and rescues the chromatin phenotype of cultured fibroblasts, including histone methylation status and BAF and LAP2alpha distribution patterns. Importantly, rapamycin treatment does not affect lamin C protein levels, but increases the relative expression of the prelamin A endoprotease ZMPSTE24. Thus, rapamycin, an antibiotic belonging to the class of macrolides, previously found to increase longevity in mouse models, can serve as a therapeutic tool, to eliminate progerin, avoid farnesylated prelamin A accumulation, and restore chromatin dynamics in progeroid laminopathies.
Figures

References
-
- Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140:2603–24. - PubMed
-
- Dominguez-Gerpe L, Araujo-Vilar D. Prematurely aged children: molecular alterations leading to Hutchinson-Gilford progeria and Werner syndromes. Curr Aging Sci. 2008;1:202–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

