Structure, fluctuation and magnitude of a natural grassland soil metagenome
- PMID: 22297556
- PMCID: PMC3498926
- DOI: 10.1038/ismej.2011.197
Structure, fluctuation and magnitude of a natural grassland soil metagenome
Abstract
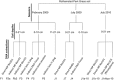
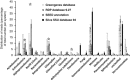
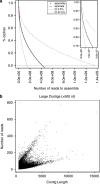
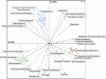
The soil ecosystem is critical for human health, affecting aspects of the environment from key agricultural and edaphic parameters to critical influence on climate change. Soil has more unknown biodiversity than any other ecosystem. We have applied diverse DNA extraction methods coupled with high throughput pyrosequencing to explore 4.88 × 10(9) bp of metagenomic sequence data from the longest continually studied soil environment (Park Grass experiment at Rothamsted Research in the UK). Results emphasize important DNA extraction biases and unexpectedly low seasonal and vertical soil metagenomic functional class variations. Clustering-based subsystems and carbohydrate metabolism had the largest quantity of annotated reads assigned although <50% of reads were assigned at an E value cutoff of 10(-5). In addition, with the more detailed subsystems, cAMP signaling in bacteria (3.24±0.27% of the annotated reads) and the Ton and Tol transport systems (1.69±0.11%) were relatively highly represented. The most highly represented genome from the database was that for a Bradyrhizobium species. The metagenomic variance created by integrating natural and methodological fluctuations represents a global picture of the Rothamsted soil metagenome that can be used for specific questions and future inter-environmental metagenomic comparisons. However, only 1% of annotated sequences correspond to already sequenced genomes at 96% similarity and E values of <10(-5), thus, considerable genomic reconstructions efforts still have to be performed.
Figures


References
-
- Agarwal N, Bishai WR. cAMP signaling in Mycobacterium tuberculosis. Indian J Exp Biol. 2009;47:393–400. - PubMed
-
- Akhter Y, Yellaboina S, Farhana A, Ranjan A, Ahmed N, Hasnain SE. Genome scale portrait of cAMP-receptor protein (CRP) regulons in mycobacteria points to their role in pathogenesis. Gene. 2008;407:148–158. - PubMed
-
- Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. - PubMed
-
- Bertrand H, Poly F, Van VT, Lombard N, Nalin R, Vogel TM, et al. High molecular weight DNA recovery from soils prerequisite for biotechnological metagenomic library construction. J Microbiol Methods. 2005;62:1–11. - PubMed
-
- Boyd PW, Jickells T, Law CS, Blain S, Boyle EA, Buesseler KO, et al. Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science. 2007;315:612–617. - PubMed

