A test for selection employing quantitative trait locus and mutation accumulation data
- PMID: 22298701
- PMCID: PMC3316662
- DOI: 10.1534/genetics.111.137075
A test for selection employing quantitative trait locus and mutation accumulation data
Abstract
Evolutionary biologists attribute much of the phenotypic diversity observed in nature to the action of natural selection. However, for many phenotypic traits, especially quantitative phenotypic traits, it has been challenging to test for the historical action of selection. An important challenge for biologists studying quantitative traits, therefore, is to distinguish between traits that have evolved under the influence of strong selection and those that have evolved neutrally. Most existing tests for selection employ molecular data, but selection also leaves a mark on the genetic architecture underlying a trait. In particular, the distribution of quantitative trait locus (QTL) effect sizes and the distribution of mutational effects together provide information regarding the history of selection. Despite the increasing availability of QTL and mutation accumulation data, such data have not yet been effectively exploited for this purpose. We present a model of the evolution of QTL and employ it to formulate a test for historical selection. To provide a baseline for neutral evolution of the trait, we estimate the distribution of mutational effects from mutation accumulation experiments. We then apply a maximum-likelihood-based method of inference to estimate the range of selection strengths under which such a distribution of mutations could generate the observed QTL. Our test thus represents the first integration of population genetic theory and QTL data to measure the historical influence of selection.
Figures
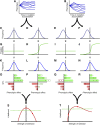
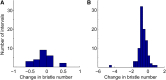
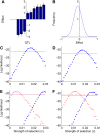
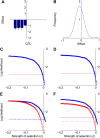

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

