Non-host defense response in a novel Arabidopsis-Xanthomonas citri subsp. citri pathosystem
- PMID: 22299054
- PMCID: PMC3267768
- DOI: 10.1371/journal.pone.0031130
Non-host defense response in a novel Arabidopsis-Xanthomonas citri subsp. citri pathosystem
Abstract
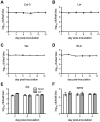
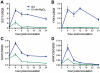
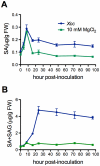
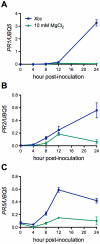
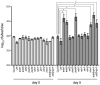
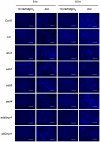
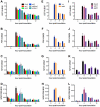
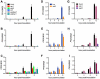
Citrus canker, caused by Xanthomonas citri subsp. citri (Xcc), is one of the most destructive diseases of citrus. Progress of breeding citrus canker-resistant varieties is modest due to limited resistant germplasm resources and lack of candidate genes for genetic manipulation. The objective of this study is to establish a novel heterologous pathosystem between Xcc and the well-established model plant Arabidopsis thaliana for defense mechanism dissection and resistance gene identification. Our results indicate that Xcc bacteria neither grow nor decline in Arabidopsis, but induce multiple defense responses including callose deposition, reactive oxygen species and salicylic aicd (SA) production, and defense gene expression, indicating that Xcc activates non-host resistance in Arabidopsis. Moreover, Xcc-induced defense gene expression is suppressed or attenuated in several well-characterized SA signaling mutants including eds1, pad4, eds5, sid2, and npr1. Interestingly, resistance to Xcc is compromised only in eds1, pad4, and eds5, but not in sid2 and npr1. However, combining sid2 and npr1 in the sid2npr1 double mutant compromises resistance to Xcc, suggesting genetic interactions likely exist between SID2 and NPR1 in the non-host resistance against Xcc in Arabidopsis. These results demonstrate that the SA signaling pathway plays a critical role in regulating non-host defense against Xcc in Arabidopsis and suggest that the SA signaling pathway genes may hold great potential for breeding citrus canker-resistant varieties through modern gene transfer technology.
Conflict of interest statement
Figures

References
-
- Gottwald TR, Graham JH, Civerolo EL, Barrett HC, Hearn CJ. Differential host range reaction of citrus and citrus relatives to citrus canker and citrus bacterial spot determined by leaf mesophyll susceptibility. Plant Dis. 1993;77:1004–1009.
-
- Graham JH, Leite RP. Lack of control of citrus canker by induced systemic resistance compounds. Plant Dis. 2004;88:745–750. - PubMed
-
- Zhang X, Francis MI, Dawson WO, Graham JH, Orbović V, et al. Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur J Plant Pathol. 2010;128:91–100.
-
- Viloria Z, Drouilard DL, Grahm JH, Grosser JW. Screening triploid hybrids of ‘Lakeland’ Limequat for resistance to citrus canker. Plant Dis. 2004;88:1056–1060. - PubMed
-
- Mishina TE, Zeier J. Bacterial non-host resistance: interaction of Arabidopsis with non-adapted Pseudomonas syringae strains. Physiol Plant. 2007;131:448–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

