Distance and helical phase dependence of synergistic transcription activation in cis-regulatory module
- PMID: 22299056
- PMCID: PMC3267773
- DOI: 10.1371/journal.pone.0031198
Distance and helical phase dependence of synergistic transcription activation in cis-regulatory module
Abstract
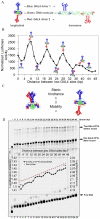
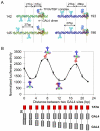
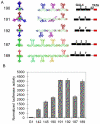
Deciphering of the spatial and stereospecific constraints on synergistic transcription activation mediated between activators bound to cis-regulatory elements is important for understanding gene regulation and remains largely unknown. It has been commonly believed that two activators will activate transcription most effectively when they are bound on the same face of DNA double helix and within a boundary distance from the transcription initiation complex attached to the TATA box. In this work, we studied the spatial and stereospecific constraints on activation by multiple copies of bound model activators using a series of engineered relative distances and stereospecific orientations. We observed that multiple copies of the activators GAL4-VP16 and ZEBRA bound to engineered promoters activated transcription more effectively when bound on opposite faces of the DNA double helix. This phenomenon was not affected by the spatial relationship between the proximal activator and initiation complex. To explain these results, we proposed the novel concentration field model, which posits the effective concentration of bound activators, and therefore the transcription activation potential, is affected by their stereospecific positioning. These results could be used to understand synergistic transcription activation anew and to aid the development of predictive models for the identification of cis-regulatory elements.
Conflict of interest statement
Figures




Similar articles
-
The ligand-binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation.Mol Cell Biol. 1995 Mar;15(3):1817-25. doi: 10.1128/MCB.15.3.1817. Mol Cell Biol. 1995. PMID: 7862171 Free PMC article.
-
Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex.Plant Cell. 1999 Aug;11(8):1591-602. doi: 10.1105/tpc.11.8.1591. Plant Cell. 1999. PMID: 10449590 Free PMC article.
-
Synergism between Tat and VP16 in trans-activation of HIV-1 LTR.J Mol Biol. 1993 Dec 5;234(3):610-9. doi: 10.1006/jmbi.1993.1615. J Mol Biol. 1993. PMID: 8254663
-
DNA constraints on transcription activation in vitro.J Mol Biol. 2000 Mar 24;297(2):321-34. doi: 10.1006/jmbi.2000.3562. J Mol Biol. 2000. PMID: 10715204
-
Mechanisms of viral activators.Cold Spring Harb Symp Quant Biol. 1998;63:243-52. doi: 10.1101/sqb.1998.63.243. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384288 Review.
Cited by
-
Parallel Reporter Assays Identify Altered Regulatory Role of rs684232 in Leading to Prostate Cancer Predisposition.Int J Mol Sci. 2021 Aug 16;22(16):8792. doi: 10.3390/ijms22168792. Int J Mol Sci. 2021. PMID: 34445492 Free PMC article.
-
Can genome engineering be used to target cancer-associated enhancers?Epigenomics. 2014;6(5):493-501. doi: 10.2217/epi.14.30. Epigenomics. 2014. PMID: 25431942 Free PMC article. Review.
-
Evolutionary Footprints of Short Tandem Repeats in Avian Promoters.Sci Rep. 2016 Jan 14;6:19421. doi: 10.1038/srep19421. Sci Rep. 2016. PMID: 26766026 Free PMC article.
-
Assembly PCR synthesis of optimally designed, compact, multi-responsive promoters suited to gene therapy application.Sci Rep. 2016 Jul 8;6:29388. doi: 10.1038/srep29388. Sci Rep. 2016. PMID: 27387837 Free PMC article.
-
Functional Screenings Identify Regulatory Variants Associated with Breast Cancer Susceptibility.Curr Issues Mol Biol. 2021 Oct 26;43(3):1756-1777. doi: 10.3390/cimb43030124. Curr Issues Mol Biol. 2021. PMID: 34889888 Free PMC article.
References
-
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

