Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation
- PMID: 22301074
- PMCID: PMC3270475
- DOI: 10.1093/database/bar068
Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation
Abstract
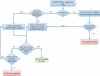
InterPro amalgamates predictive protein signatures from a number of well-known partner databases into a single resource. To aid with interpretation of results, InterPro entries are manually annotated with terms from the Gene Ontology (GO). The InterPro2GO mappings are comprised of the cross-references between these two resources and are the largest source of GO annotation predictions for proteins. Here, we describe the protocol by which InterPro curators integrate GO terms into the InterPro database. We discuss the unique challenges involved in integrating specific GO terms with entries that may describe a diverse set of proteins, and we illustrate, with examples, how InterPro hierarchies reflect GO terms of increasing specificity. We describe a revised protocol for GO mapping that enables us to assign GO terms to domains based on the function of the individual domain, rather than the function of the families in which the domain is found. We also discuss how taxonomic constraints are dealt with and those cases where we are unable to add any appropriate GO terms. Expert manual annotation of InterPro entries with GO terms enables users to infer function, process or subcellular information for uncharacterized sequences based on sequence matches to predictive models. Database URL: http://www.ebi.ac.uk/interpro. The complete InterPro2GO mappings are available at: ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/external2go/interpro2go.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

