HIV-1 capsid-targeting domain of cleavage and polyadenylation specificity factor 6
- PMID: 22301135
- PMCID: PMC3302544
- DOI: 10.1128/JVI.06607-11
HIV-1 capsid-targeting domain of cleavage and polyadenylation specificity factor 6
Abstract
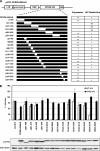
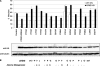
The antiviral factor CPSF6-358 restricts human immunodeficiency virus type 1 (HIV-1) infection through an interaction with capsid (CA), preventing virus nuclear entry and integration. HIV-1 acquires resistance to CPSF6-358 through an N74D mutation of CA that impairs binding of the antiviral factor. Here we examined the determinants within CPSF6-358 that are necessary for CA-specific interaction. Residues 314 to 322 include amino acids that are essential for CPSF6-358 restriction of HIV-1. Fusion of CPSF6 residues 301 to 358 to rhesus TRIM5α is also sufficient to restrict wild-type but not N74D HIV-1. Restriction is lost if CPSF6 residues in the amino acid 314 to 322 interaction motif are mutated. Examination of the CA targeting motif in CPSF6-358 did not reveal evidence of positive selection. Given the sensitivity of different primate lentiviruses to CPSF6-358 and apparent conservation of this interaction, our data suggest that CPSF6-358-mediated targeting of HIV-1 could provide a broadly effective antiviral strategy.
Figures



Similar articles
-
HIV-1 is more dependent on the K182 capsid residue than HIV-2 for interactions with CPSF6.Virology. 2019 Jun;532:118-126. doi: 10.1016/j.virol.2019.04.012. Epub 2019 Apr 27. Virology. 2019. PMID: 31071616
-
Truncated CPSF6 Forms Higher-Order Complexes That Bind and Disrupt HIV-1 Capsid.J Virol. 2018 Jun 13;92(13):e00368-18. doi: 10.1128/JVI.00368-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29643241 Free PMC article.
-
The Cleavage and Polyadenylation Specificity Factor 6 (CPSF6) Subunit of the Capsid-recruited Pre-messenger RNA Cleavage Factor I (CFIm) Complex Mediates HIV-1 Integration into Genes.J Biol Chem. 2016 May 27;291(22):11809-19. doi: 10.1074/jbc.M116.721647. Epub 2016 Mar 18. J Biol Chem. 2016. PMID: 26994143 Free PMC article.
-
HIV-1-induced translocation of CPSF6 to biomolecular condensates.Trends Microbiol. 2024 Aug;32(8):781-790. doi: 10.1016/j.tim.2024.01.001. Epub 2024 Jan 23. Trends Microbiol. 2024. PMID: 38267295 Free PMC article. Review.
-
HIV Capsid and Integration Targeting.Viruses. 2021 Jan 18;13(1):125. doi: 10.3390/v13010125. Viruses. 2021. PMID: 33477441 Free PMC article. Review.
Cited by
-
Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core.J Mol Biol. 2013 Dec 13;425(24):5032-44. doi: 10.1016/j.jmb.2013.07.025. Epub 2013 Jul 23. J Mol Biol. 2013. PMID: 23886867 Free PMC article.
-
Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):18625-30. doi: 10.1073/pnas.1419945112. Epub 2014 Dec 17. Proc Natl Acad Sci U S A. 2014. PMID: 25518861 Free PMC article.
-
Extreme genetic fragility of the HIV-1 capsid.PLoS Pathog. 2013;9(6):e1003461. doi: 10.1371/journal.ppat.1003461. Epub 2013 Jun 20. PLoS Pathog. 2013. PMID: 23818857 Free PMC article.
-
CPSF6 promotes HIV-1 preintegration complex function.J Virol. 2025 May 20;99(5):e0049025. doi: 10.1128/jvi.00490-25. Epub 2025 Apr 9. J Virol. 2025. PMID: 40202316 Free PMC article.
-
Human Immunodeficiency Virus 1 (HIV-1): Viral Latency, the Reservoir, and the Cure.Yale J Biol Med. 2020 Sep 30;93(4):549-560. eCollection 2020 Sep. Yale J Biol Med. 2020. PMID: 33005119 Free PMC article. Review.
References
-
- Bartz SR, Vodicka MA. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337–342 - PubMed
-
- Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

