Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection
- PMID: 22301145
- PMCID: PMC3318671
- DOI: 10.1128/JVI.05887-11
Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection
Abstract
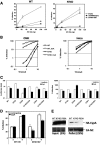
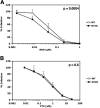
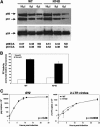
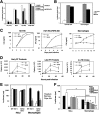
The antiviral factor CPSF6-358 interferes with the nuclear entry of human immunodeficiency virus type 1 (HIV-1). HIV-1 acquires resistance to CPSF6-358 through the N74D mutation of the capsid (CA), which alters its nuclear entry pathway. Here we show that compared to wild-type (WT) HIV-1, N74D HIV-1 is more sensitive to cyclosporine, has increased sensitivity to nevirapine, and is impaired in macrophage infection prior to reverse transcription. These phenotypes suggest a difference in the N74D reverse transcription complex that manifests early after infection and prior to interaction with the nuclear pore. Overall, our data indicate that N74D HIV-1 replication in transformed cells requires cyclophilin A but is dependent on other interactions in macrophages.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

