A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice
- PMID: 22301384
- PMCID: PMC3346221
- DOI: 10.1093/jxb/err431
A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice
Abstract
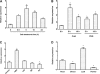
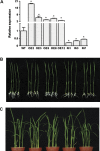
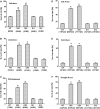
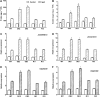
MYB-type transcription factors play a diverse role in plant development and response to abiotic stress. This study isolated a rice R2R3-type MYB gene, OsMYB2, and functionally characterized its role in tolerance to abiotic stress by generating transgenic rice plants with overexpressing and RNA interference OsMYB2. Expression of OsMYB2 was up-regulated by salt, cold, and dehydration stress. OsMYB2 was localized in the nucleus with transactivation activity. No difference in growth and development between the OsMYB2-overexpressing and wild-type plants was observed under normal growth conditions, but the OsMYB2-overexpressing plants were more tolerant to salt, cold, and dehydration stresses and more sensitive to abscisic acid than wild-type plants. The OsMYB2-overexpressing plants accumulated greater amounts of soluble sugars and proline than wild-type plants under salt stress. Overexpression of OsMYB2 enhanced up-regulation of genes encoding proline synthase and transporters. The OsMYB2-overexpressing plants accumulated less amounts of H(2)O(2) and malondialdehyde. The enhanced activities of antioxidant enzymes, including peroxidase, superoxide dismutase, and catalase, may underlie the lower H(2)O(2) contents in OsMYB2-overexpressing plants. There was greater up-regulation of stress-related genes, including OsLEA3, OsRab16A, and OsDREB2A, in the OsMYB2-overexpressing plants. Microarray analysis showed that expression of numerous genes involving diverse functions in stress response was altered in the OsMYB2-overexpressing plants. These findings suggest that OsMYB2 encodes a stress-responsive MYB transcription factor that plays a regulatory role in tolerance of rice to salt, cold, and dehydration stress.
Figures





Similar articles
-
Genome-wide identification of R2R3-MYB transcription factor subfamily genes involved in salt stress in rice (Oryza sativa L.).BMC Genomics. 2024 Aug 23;25(1):797. doi: 10.1186/s12864-024-10693-5. BMC Genomics. 2024. PMID: 39179980 Free PMC article.
-
Transcription factor OsMYB2 triggers amino acid transporter OsANT1 expression to regulate rice growth and salt tolerance.Plant Physiol. 2025 Feb 7;197(2):kiae559. doi: 10.1093/plphys/kiae559. Plant Physiol. 2025. PMID: 39425973 Free PMC article.
-
Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress.Planta. 2011 Aug;234(2):331-45. doi: 10.1007/s00425-011-1403-2. Epub 2011 Mar 30. Planta. 2011. PMID: 21448719
-
A Meta-Analysis of Comparative Transcriptomic Data Reveals a Set of Key Genes Involved in the Tolerance to Abiotic Stresses in Rice.Int J Mol Sci. 2019 Nov 12;20(22):5662. doi: 10.3390/ijms20225662. Int J Mol Sci. 2019. PMID: 31726733 Free PMC article.
-
The network centered on ICEs play roles in plant cold tolerance, growth and development.Planta. 2022 Mar 6;255(4):81. doi: 10.1007/s00425-022-03858-7. Planta. 2022. PMID: 35249133 Review.
Cited by
-
MYB transcription factors, their regulation and interactions with non-coding RNAs during drought stress in Brassica juncea.BMC Plant Biol. 2024 Oct 24;24(1):999. doi: 10.1186/s12870-024-05736-8. BMC Plant Biol. 2024. PMID: 39448923 Free PMC article.
-
CsMYB Transcription Factors Participate in Jasmonic Acid Signal Transduction in Response to Cold Stress in Tea Plant (Camellia sinensis).Plants (Basel). 2022 Oct 27;11(21):2869. doi: 10.3390/plants11212869. Plants (Basel). 2022. PMID: 36365324 Free PMC article.
-
Transcriptome analysis of MYB transcription factors family and PgMYB genes involved in salt stress resistance in Panax ginseng.BMC Plant Biol. 2022 Oct 8;22(1):479. doi: 10.1186/s12870-022-03871-8. BMC Plant Biol. 2022. PMID: 36209052 Free PMC article.
-
Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice.Sci Rep. 2021 Mar 4;11(1):5166. doi: 10.1038/s41598-021-84638-3. Sci Rep. 2021. PMID: 33664392 Free PMC article.
-
Ectopic Expression of a Wheat R2R3-Type MYB Gene in Transgenic Tobacco Enhances Osmotic Stress Tolerance via Maintaining ROS Balance and Improving Root System Architecture.Biology (Basel). 2024 Feb 18;13(2):128. doi: 10.3390/biology13020128. Biology (Basel). 2024. PMID: 38392346 Free PMC article.
References
-
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. - PubMed
-
- Alexieva V, Sergiev I, Mapelli S, Karanov E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant, Cell and Environment. 2001;24:1337–1344.
-
- Armengaud P, Thiery L, Buhot N, Grenier-De March G, Savoure A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiologia Plantarum. 2004;120:442–450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

