Analysis of high accuracy, quantitative proteomics data in the MaxQB database
- PMID: 22301388
- PMCID: PMC3316731
- DOI: 10.1074/mcp.M111.014068
Analysis of high accuracy, quantitative proteomics data in the MaxQB database
Abstract
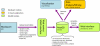
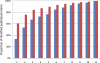
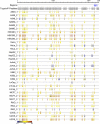
MS-based proteomics generates rapidly increasing amounts of precise and quantitative information. Analysis of individual proteomic experiments has made great strides, but the crucial ability to compare and store information across different proteome measurements still presents many challenges. For example, it has been difficult to avoid contamination of databases with low quality peptide identifications, to control for the inflation in false positive identifications when combining data sets, and to integrate quantitative data. Although, for example, the contamination with low quality identifications has been addressed by joint analysis of deposited raw data in some public repositories, we reasoned that there should be a role for a database specifically designed for high resolution and quantitative data. Here we describe a novel database termed MaxQB that stores and displays collections of large proteomics projects and allows joint analysis and comparison. We demonstrate the analysis tools of MaxQB using proteome data of 11 different human cell lines and 28 mouse tissues. The database-wide false discovery rate is controlled by adjusting the project specific cutoff scores for the combined data sets. The 11 cell line proteomes together identify proteins expressed from more than half of all human genes. For each protein of interest, expression levels estimated by label-free quantification can be visualized across the cell lines. Similarly, the expression rank order and estimated amount of each protein within each proteome are plotted. We used MaxQB to calculate the signal reproducibility of the detected peptides for the same proteins across different proteomes. Spearman rank correlation between peptide intensity and detection probability of identified proteins was greater than 0.8 for 64% of the proteome, whereas a minority of proteins have negative correlation. This information can be used to pinpoint false protein identifications, independently of peptide database scores. The information contained in MaxQB, including high resolution fragment spectra, is accessible to the community via a user-friendly web interface at http://www.biochem.mpg.de/maxqb.
Figures




References
-
- Mallick P., Kuster B. (2010) Proteomics: A pragmatic perspective. Nat. Biotechnol. 28, 695–709 - PubMed
-
- Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 - PubMed
-
- Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 - PubMed
-
- Olsen J. V., Mann M. (2011) Effective representation and storage of mass spectrometry-based proteomic data sets for the scientific community. Sci. Signal. 4, pe7. - PubMed
-
- Taylor C. F., Paton N. W., Garwood K. L., Kirby P. D., Stead D. A., Yin Z., Deutsch E. W., Selway L., Walker J., Riba-Garcia I., Mohammed S., Deery M. J., Howard J. A., Dunkley T., Aebersold R., Kell D. B., Lilley K. S., Roepstorff P., Yates J. R., 3rd, Brass A., Brown A. J., Cash P., Gaskell S. J., Hubbard S. J., Oliver S. G. (2003) A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 21, 247–254 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

