LRRK2 inhibition attenuates microglial inflammatory responses
- PMID: 22302802
- PMCID: PMC3532034
- DOI: 10.1523/JNEUROSCI.5601-11.2012
LRRK2 inhibition attenuates microglial inflammatory responses
Erratum in
-
Erratum: Moehle et al., "LRRK2 Inhibition Attenuates Microglial Inflammatory Responses".J Neurosci. 2022 Feb 2;42(5):938. doi: 10.1523/JNEUROSCI.2507-21.2021. Epub 2022 Jan 26. J Neurosci. 2022. PMID: 35082121 Free PMC article. No abstract available.
Abstract
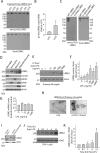
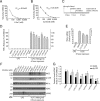
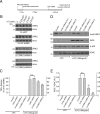
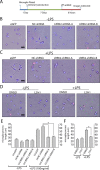
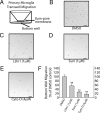
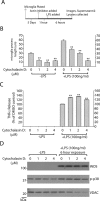
Missense mutations in leucine-rich repeat kinase 2 (LRRK2) cause late-onset Parkinson's disease (PD), and common genetic variation in LRRK2 modifies susceptibility to Crohn's disease and leprosy. High levels of LRRK2 expression in peripheral monocytes and macrophages suggest a role for LRRK2 in these cells, yet little is known about LRRK2 expression and function in immune cells of the brain. Here, we demonstrate a role for LRRK2 in mediating microglial proinflammatory responses and morphology. In a murine model of neuroinflammation, we observe robust induction of LRRK2 in microglia. Experiments with toll-like receptor 4 (TLR4)-stimulated rat primary microglia show that inflammation increases LRRK2 activity and expression, while inhibition of LRRK2 kinase activity or knockdown of protein attenuates TNFα secretion and nitric oxide synthase (iNOS) induction. LRRK2 inhibition blocks TLR4 stimulated microglial process outgrowth and impairs ADP stimulated microglial chemotaxis. However, actin inhibitors that phenocopy inhibition of process outgrowth and chemotaxis fail to modify TLR4 stimulation of TNFα secretion and inducible iNOS induction, suggesting that LRRK2 acts upstream of cytoskeleton control as a stress-responsive kinase. These data demonstrate LRRK2 in regulating responses in immune cells of the brain and further implicate microglial involvement in late-onset PD.
Figures


References
-
- Balcaitis S, Weinstein JR, Li S, Chamberlain JS, Möller T. Lentiviral transduction of microglial cells. Glia. 2005;50:48–55. - PubMed
-
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, Faull RL, Emson PC, Torp R, Ottersen OP, Dawson TM, Dawson VL. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. - PubMed
-
- Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. - PubMed
-
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
