Assembly, purification, and pre-steady-state kinetic analysis of active RNA-dependent RNA polymerase elongation complex
- PMID: 22303022
- PMCID: PMC3323022
- DOI: 10.1074/jbc.M111.325530
Assembly, purification, and pre-steady-state kinetic analysis of active RNA-dependent RNA polymerase elongation complex
Abstract
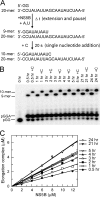
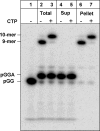
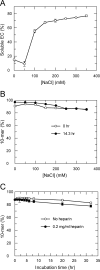
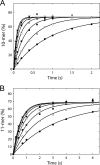
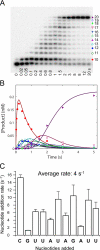
NS5B is the RNA-dependent RNA polymerase responsible for replicating hepatitis C virus (HCV) genomic RNA. Despite more than a decade of work, the formation of a highly active NS5B polymerase·RNA complex suitable for mechanistic and structural studies has remained elusive. Here, we report that through a novel way of optimizing initiation conditions, we were able to generate a productive NS5B·primer·template elongation complex stalled after formation of a 9-nucleotide primer. In contrast to previous reports of very low proportions of active NS5B, we observed that under optimized conditions up to 65% of NS5B could be converted into active elongation complexes. The elongation complex was extremely stable, allowing purification away from excess nucleotide and abortive initiation products so that the purified complex was suitable for pre-steady-state kinetic analyses of polymerase activity. Single turnover kinetic studies showed that CTP is incorporated with apparent K(d) and k(pol) values of 39 ± 3 μM and 16 ± 1 s(-1), respectively, giving a specificity constant of k(pol)/K(d) of 0.41 μM(-1) s(-1). The kinetics of multiple nucleotide incorporation during processive elongation also were determined. This work establishes a novel way to generate a highly active elongation complex of the medically important NS5B polymerase for structural and functional studies.
Figures
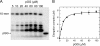
Similar articles
-
Specific inhibitors of HCV polymerase identified using an NS5B with lower affinity for template/primer substrate.Nucleic Acids Res. 2004 Jan 22;32(2):422-31. doi: 10.1093/nar/gkh160. Print 2004. Nucleic Acids Res. 2004. PMID: 14739234 Free PMC article.
-
Functional characterization of fingers subdomain-specific monoclonal antibodies inhibiting the hepatitis C virus RNA-dependent RNA polymerase.J Biol Chem. 2008 Aug 29;283(35):24089-102. doi: 10.1074/jbc.M803422200. Epub 2008 Jun 23. J Biol Chem. 2008. PMID: 18574240 Free PMC article.
-
Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation.J Biol Chem. 2008 Dec 5;283(49):33893-901. doi: 10.1074/jbc.M803094200. Epub 2008 Oct 6. J Biol Chem. 2008. PMID: 18840605 Free PMC article.
-
Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus.J Viral Hepat. 2000 May;7(3):167-74. doi: 10.1046/j.1365-2893.2000.00218.x. J Viral Hepat. 2000. PMID: 10849258 Review.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
Cited by
-
Kinetics of elementary steps in loop-mediated isothermal amplification (LAMP) show that strand invasion during initiation is rate-limiting.Nucleic Acids Res. 2023 Jan 11;51(1):488-499. doi: 10.1093/nar/gkac1221. Nucleic Acids Res. 2023. PMID: 36583345 Free PMC article.
-
Evolving Diversity of Hepatitis C Viruses in Yunnan Honghe, China.Int J Mol Sci. 2016 Mar 18;17(3):403. doi: 10.3390/ijms17030403. Int J Mol Sci. 2016. PMID: 26999127 Free PMC article.
-
Cystoviral RNA-directed RNA polymerases: Regulation of RNA synthesis on multiple time and length scales.Virus Res. 2017 Apr 15;234:135-152. doi: 10.1016/j.virusres.2017.01.006. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104452 Free PMC article. Review.
-
Crystal structure of a tick-borne flavivirus RNA-dependent RNA polymerase suggests a host adaptation hotspot in RNA viruses.Nucleic Acids Res. 2021 Feb 22;49(3):1567-1580. doi: 10.1093/nar/gkaa1250. Nucleic Acids Res. 2021. PMID: 33406260 Free PMC article.
-
Rate-limiting pyrophosphate release by hepatitis C virus polymerase NS5B improves fidelity.J Biol Chem. 2020 Nov 27;295(48):16436-16444. doi: 10.1074/jbc.RA120.015394. Epub 2020 Sep 16. J Biol Chem. 2020. PMID: 32938715 Free PMC article.
References
-
- Tellinghuisen T. L. (2011) An overview of the hepatitis C virus life cycle in Hepatitis C: Antiviral Drug Discovery and Development (Tan S.-L., He Y., eds), pp. 1–48, Caister Academic Press, Norfolk, UK
-
- Dutartre H., Boretto J., Guillemot J. C., Canard B. (2005) A relaxed discrimination of 2′-O-methyl-GTP relative to GTP between de novo and Elongative RNA synthesis by the hepatitis C RNA-dependent RNA polymerase NS5B. J. Biol. Chem. 280, 6359–6368 - PubMed
-
- Cramer J., Jaeger J., Restle T. (2006) Biochemical and pre-steady-state kinetic characterization of the hepatitis C virus RNA polymerase (NS5BΔ21, HC-J4). Biochemistry 45, 3610–3619 - PubMed
-
- Selisko B., Dutartre H., Guillemot J. C., Debarnot C., Benarroch D., Khromykh A., Desprès P., Egloff M. P., Canard B. (2006) Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351, 145–158 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

