Systematic Curation of miRBase Annotation Using Integrated Small RNA High-Throughput Sequencing Data for C. elegans and Drosophila
- PMID: 22303321
- PMCID: PMC3268580
- DOI: 10.3389/fgene.2011.00025
Systematic Curation of miRBase Annotation Using Integrated Small RNA High-Throughput Sequencing Data for C. elegans and Drosophila
Abstract
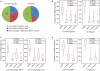
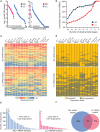
MicroRNAs (miRNAs) are a class of 20-23 nucleotide small RNAs that regulate gene expression post-transcriptionally in animals and plants. Annotation of miRNAs by the miRNA database (miRBase) has largely relied on computational approaches. As a result, many miRBase entries lack experimental validation, and discrepancies between miRBase annotation and actual miRNA sequences are often observed. In this study, we integrated the small RNA sequencing (smRNA-seq) datasets in Caenorhabditis elegans and Drosophila melanogaster and devised an analytical pipeline coupled with detailed manual inspection to curate miRNA annotation systematically in miRBase. Our analysis reveals 19 (17.0%) and 51 (31.3%) miRNAs entries with detectable smRNA-seq reads have mature sequence discrepancies in C. elegans and D. melanogaster, respectively. These discrepancies frequently occur either for conserved miRNA families whose mature sequences were predicted according to their homologous counterparts in other species or for miRNAs whose precursor miRNA (pre-miRNA) hairpins produce an abundance of multiple miRNA isoforms or variants. Our analysis shows that while Drosophila pre-miRNAs, on average, produce less than 60% accurate mature miRNA reads in addition to their 5' and 3' variant isoforms, the precision of miRNA processing in C. elegans is much higher, at over 90%. Based on the revised miRNA sequences, we analyzed expression patterns of the more conserved (MC) and less conserved (LC) miRNAs and found that, whereas MC miRNAs are often co-expressed at multiple developmental stages, LC miRNAs tend to be expressed specifically at fewer stages.
Keywords: database curation; deep sequencing; microRNA.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

