Systems Biology Reveals MicroRNA-Mediated Gene Regulation
- PMID: 22303325
- PMCID: PMC3268584
- DOI: 10.3389/fgene.2011.00029
Systems Biology Reveals MicroRNA-Mediated Gene Regulation
Abstract
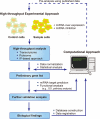
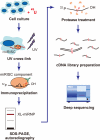
MicroRNAs (miRNAs) are members of the small non-coding RNAs, which are principally known for their functions as post-transcriptional regulators of target genes. Regulation by miRNAs is triggered by the translational repression or degradation of their complementary target messenger RNAs (mRNAs). The growing number of reported miRNAs and the estimate that hundreds or thousands of genes are regulated by them suggest a magnificent gene regulatory network in which these molecules are embedded. Indeed, recent reports have suggested critical roles for miRNAs in various biological functions, such as cell differentiation, development, oncogenesis, and the immune responses, which are mediated by systems-wide changes in gene expression profiles. Therefore, it is essential to analyze this complex regulatory network at the transcriptome and proteome levels, which should be possible with approaches that include both high-throughput experiments and computational methodologies. Here, we introduce several systems-level approaches that have been applied to miRNA research, and discuss their potential to reveal miRNA-guided gene regulatory systems and their impacts on biological functions.
Keywords: gene regulatory network; immunoprecipitation; microRNA; proteome; systems biology; transcriptome.
Figures

Similar articles
-
Integrated analyses to reconstruct microRNA-mediated regulatory networks in mouse liver using high-throughput profiling.BMC Genomics. 2015;16 Suppl 2(Suppl 2):S12. doi: 10.1186/1471-2164-16-S2-S12. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25707768 Free PMC article.
-
MicroRNA expression and gene regulation drive breast cancer progression and metastasis in PyMT mice.Breast Cancer Res. 2016 Jul 22;18(1):75. doi: 10.1186/s13058-016-0735-z. Breast Cancer Res. 2016. PMID: 27449149 Free PMC article.
-
An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma.BMC Med Genomics. 2012 Jan 19;5:3. doi: 10.1186/1755-8794-5-3. BMC Med Genomics. 2012. PMID: 22260379 Free PMC article.
-
MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions.Int J Mol Sci. 2016 Oct 13;17(10):1712. doi: 10.3390/ijms17101712. Int J Mol Sci. 2016. PMID: 27754357 Free PMC article. Review.
-
Computational methods for microRNA target prediction.Methods Enzymol. 2007;427:65-86. doi: 10.1016/S0076-6879(07)27004-1. Methods Enzymol. 2007. PMID: 17720479 Review.
Cited by
-
miRNAs in Tuberculosis: New Avenues for Diagnosis and Host-Directed Therapy.Front Microbiol. 2018 Mar 29;9:602. doi: 10.3389/fmicb.2018.00602. eCollection 2018. Front Microbiol. 2018. PMID: 29651283 Free PMC article. Review.
-
microRNAs in Mycobacterial Infection: Modulation of Host Immune Response and Apoptotic Pathways.Immune Netw. 2019 Sep 18;19(5):e30. doi: 10.4110/in.2019.19.e30. eCollection 2019 Oct. Immune Netw. 2019. PMID: 31720041 Free PMC article. Review.
-
Managing Pancreatic Adenocarcinoma: A Special Focus in MicroRNA Gene Therapy.Int J Mol Sci. 2016 May 13;17(5):718. doi: 10.3390/ijms17050718. Int J Mol Sci. 2016. PMID: 27187371 Free PMC article. Review.
-
The emerging paradigm of network medicine in the study of human disease.Circ Res. 2012 Jul 20;111(3):359-74. doi: 10.1161/CIRCRESAHA.111.258541. Circ Res. 2012. PMID: 22821909 Free PMC article. Review.
-
Recent progress using systems biology approaches to better understand molecular mechanisms of immunity.Semin Immunol. 2013 Oct 31;25(3):201-8. doi: 10.1016/j.smim.2012.11.002. Epub 2012 Dec 11. Semin Immunol. 2013. PMID: 23238271 Free PMC article. Review.
References
-
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000). Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–2910.1038/75556 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

