Clusters of adaptive evolution in the human genome
- PMID: 22303346
- PMCID: PMC3268603
- DOI: 10.3389/fgene.2011.00050
Clusters of adaptive evolution in the human genome
Abstract
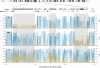
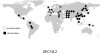
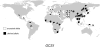
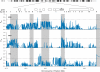
Considerable work has been devoted to identifying regions of the human genome that have been subjected to recent positive selection. Although detailed follow-up studies of putatively selected regions are critical for a deeper understanding of human evolutionary history, such studies have received comparably less attention. Recently, we have shown that ALMS1 has been the target of recent positive selection acting on standing variation in Eurasian populations. Here, we describe a careful follow-up analysis of genetic variation across the ALMS1 region, which unexpectedly revealed a cluster of substrates of positive selection. Specifically, through the analysis of SNP data from the HapMap and Human Genome Diversity Project-Centre d'Etude du Polymorphisme Humain samples as well sequence data from the region, we find compelling evidence for three independent and distinct signals of recent positive selection across this 3 Mb region surrounding ALMS1. Moreover, we analyzed the HapMap data to identify other putative clusters of independent selective events and conservatively discovered 19 additional clusters of adaptive evolution. This work has important implications for the interpretation of genome-scans for positive selection in humans and more broadly contributes to a better understanding of how recent positive selection has shaped genetic variation across the human genome.
Keywords: clustered adaptive events.
Figures


Similar articles
-
Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history.Mol Biol Evol. 2009 Jun;26(6):1357-67. doi: 10.1093/molbev/msp045. Epub 2009 Mar 11. Mol Biol Evol. 2009. PMID: 19279085 Free PMC article.
-
Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations.Immunogenetics. 2012 Oct;64(10):719-37. doi: 10.1007/s00251-012-0629-x. Epub 2012 Jul 1. Immunogenetics. 2012. PMID: 22752190 Free PMC article.
-
SNP@Evolution: a hierarchical database of positive selection on the human genome.BMC Evol Biol. 2009 Sep 5;9:221. doi: 10.1186/1471-2148-9-221. BMC Evol Biol. 2009. PMID: 19732458 Free PMC article.
-
Identifying adaptive alleles in the human genome: from selection mapping to functional validation.Hum Genet. 2021 Feb;140(2):241-276. doi: 10.1007/s00439-020-02206-7. Epub 2020 Jul 29. Hum Genet. 2021. PMID: 32728809 Review.
-
Positive selection in the human genome: from genome scans to biological significance.Annu Rev Genomics Hum Genet. 2008;9:143-60. doi: 10.1146/annurev.genom.9.081307.164411. Annu Rev Genomics Hum Genet. 2008. PMID: 18505377 Review.
Cited by
-
Positive selection in the chromosome 16 VKORC1 genomic region has contributed to the variability of anticoagulant response in humans.PLoS One. 2012;7(12):e53049. doi: 10.1371/journal.pone.0053049. Epub 2012 Dec 28. PLoS One. 2012. PMID: 23285254 Free PMC article.
-
Measured, modeled, and causal conceptions of fitness.Front Genet. 2012 Oct 23;3:196. doi: 10.3389/fgene.2012.00196. eCollection 2012. Front Genet. 2012. PMID: 23112804 Free PMC article.
-
Recent human adaptation: genomic approaches, interpretation and insights.Nat Rev Genet. 2013 Oct;14(10):692-702. doi: 10.1038/nrg3604. Nat Rev Genet. 2013. PMID: 24052086 Free PMC article. Review.
-
Using the Coriell Personalized Medicine Collaborative Data to conduct a genome-wide association study of sleep duration.Am J Med Genet B Neuropsychiatr Genet. 2015 Dec;168(8):697-705. doi: 10.1002/ajmg.b.32362. Epub 2015 Sep 3. Am J Med Genet B Neuropsychiatr Genet. 2015. PMID: 26333835 Free PMC article.
References
-
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

