Potential biomarkers in psychiatry: focus on the cholesterol system
- PMID: 22304330
- PMCID: PMC3823072
- DOI: 10.1111/j.1582-4934.2012.01543.x
Potential biomarkers in psychiatry: focus on the cholesterol system
Abstract
Measuring biomarkers to identify and assess illness is a strategy growing in popularity and relevance. Although already in clinical use for treating and predicting cancer, no biological measurement is used clinically for any psychiatric disorder. Biomarkers could predict the course of a medical problem, and aid in determining how and when to treat. Several studies have indicated that of candidate psychiatric biomarkers detected using proteomic techniques, cholesterol and associated proteins, specifically apolipoproteins (Apos), may be of interest. Cholesterol is necessary for brain development and its synthesis continues at a lower rate in the adult brain. Apos are the protein component of lipoproteins responsible for lipid transport. There is extensive evidence that the levels of cholesterol and Apos may be disturbed in psychiatric disorders, including autistic spectrum disorders (ASD). Here, we describe putative serum biomarkers for psychiatric disorders, and the role of cholesterol and Apos in central nervous system (CNS) disorders.
© 2012 The Authors Journal of Cellular and Molecular Medicine © 2012 Foundation for Cellular and Molecular Medicine/Blackwell Publishing Ltd.
Figures

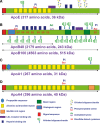
References
-
- Ross JS. Biomarker-based selection of therapy for colorectal cancer. Biomark Med. 2011;5:319–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

